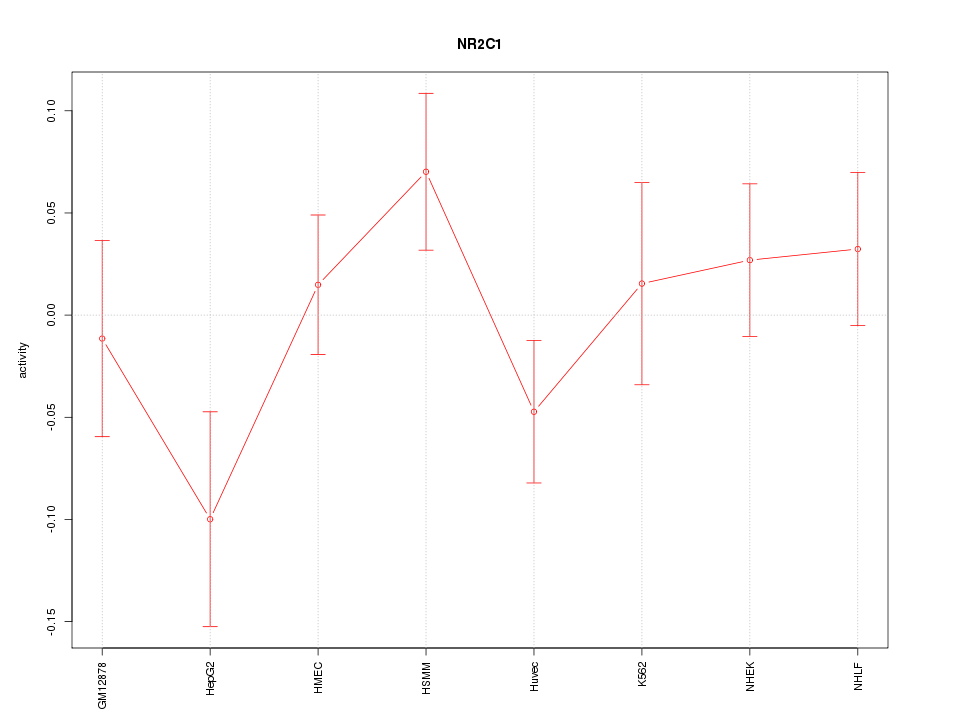
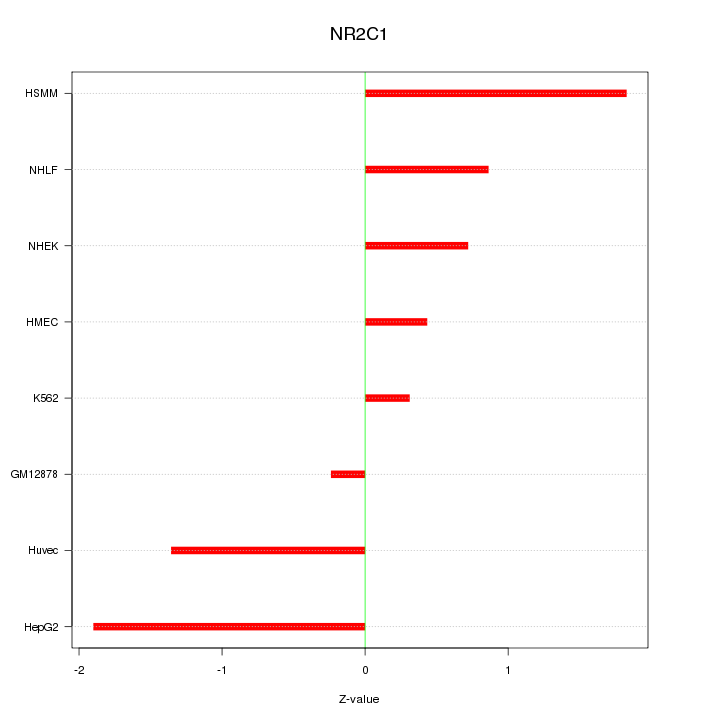
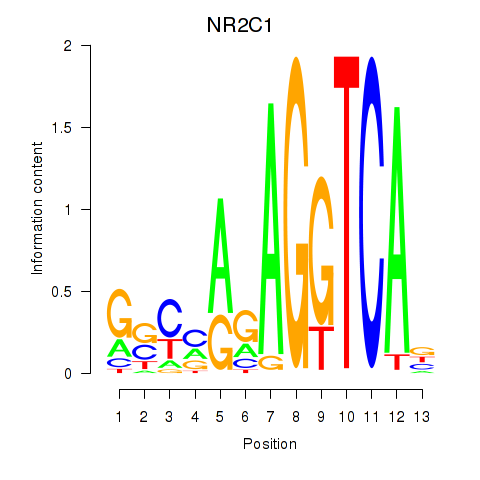
Motif ID: NR2C1
Z-value: 1.140
Transcription factors associated with NR2C1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NR2C1 | ENSG00000120798.12 | NR2C1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.3 | 1.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.3 | 1.1 | GO:0043501 | skeletal muscle adaptation(GO:0043501) |
| 0.2 | 0.8 | GO:0042335 | cuticle development(GO:0042335) |
| 0.2 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 0.6 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 1.5 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.7 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.2 | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.1 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 2.2 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.1 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.5 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 2.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0048739 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) cardiac muscle fiber development(GO:0048739) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0008356 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) asymmetric cell division(GO:0008356) |
| 0.0 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.5 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.0 | 1.3 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 2.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 2.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 2.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 1.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 2.8 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 3.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |