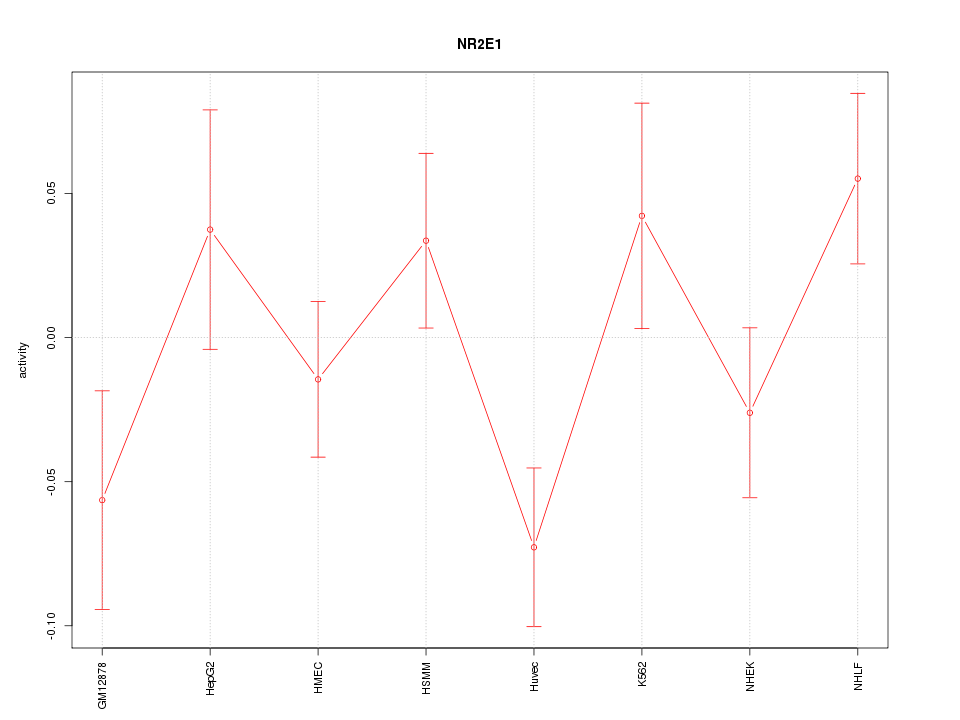
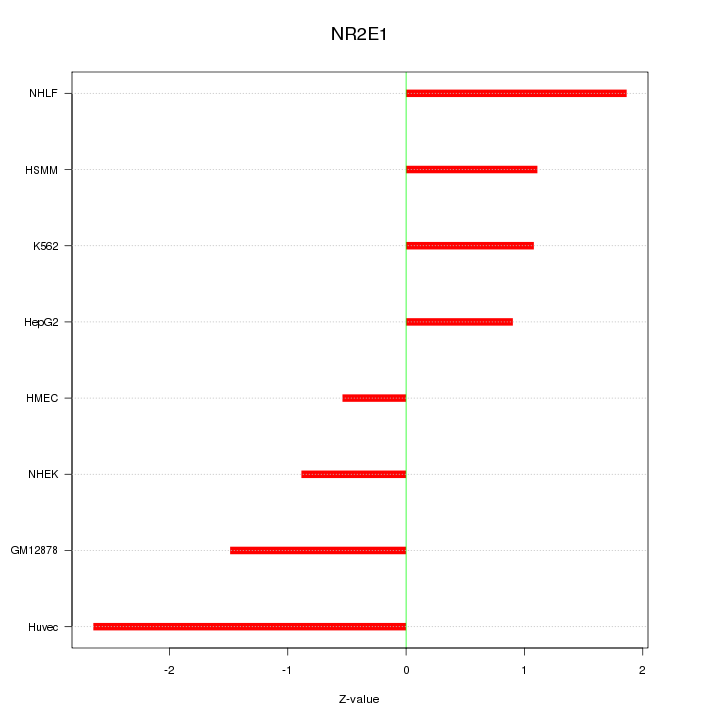
Motif ID: NR2E1
Z-value: 1.456
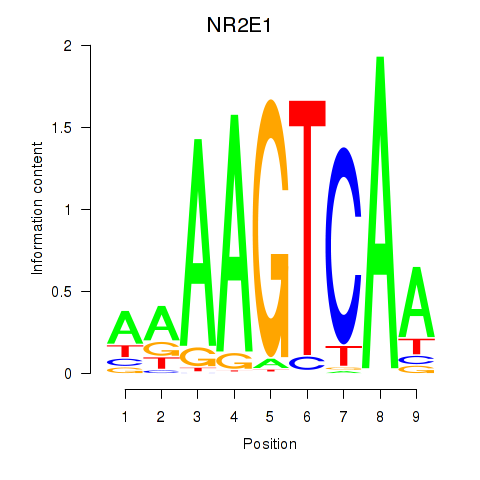
Transcription factors associated with NR2E1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NR2E1 | ENSG00000112333.7 | NR2E1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 1.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.4 | 2.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 0.7 | GO:0042759 | fatty acid elongation, saturated fatty acid(GO:0019367) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 0.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 0.5 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 1.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.7 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) regulation of cholesterol transporter activity(GO:0060694) |
| 0.1 | 0.6 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.4 | GO:0014831 | intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.1 | 0.5 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.9 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.1 | 0.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 4.3 | GO:0050931 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.8 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 0.1 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.1 | 0.2 | GO:0003289 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.4 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.8 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.5 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 1.0 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.2 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 | 0.4 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 1.3 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.5 | GO:0019054 | modulation by virus of host process(GO:0019054) modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 3.2 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.3 | GO:0060100 | regulation of phagocytosis, engulfment(GO:0060099) positive regulation of phagocytosis, engulfment(GO:0060100) regulation of membrane invagination(GO:1905153) positive regulation of membrane invagination(GO:1905155) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.3 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.3 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.4 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.4 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.6 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.0 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0030578 | nuclear body organization(GO:0030575) PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 4.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 0.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 1.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.4 | GO:0042589 | phagocytic vesicle membrane(GO:0030670) zymogen granule membrane(GO:0042589) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 2.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 8.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.6 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.1 | 0.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.1 | 3.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.7 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.5 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.6 | GO:0008194 | UDP-glycosyltransferase activity(GO:0008194) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.5 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |