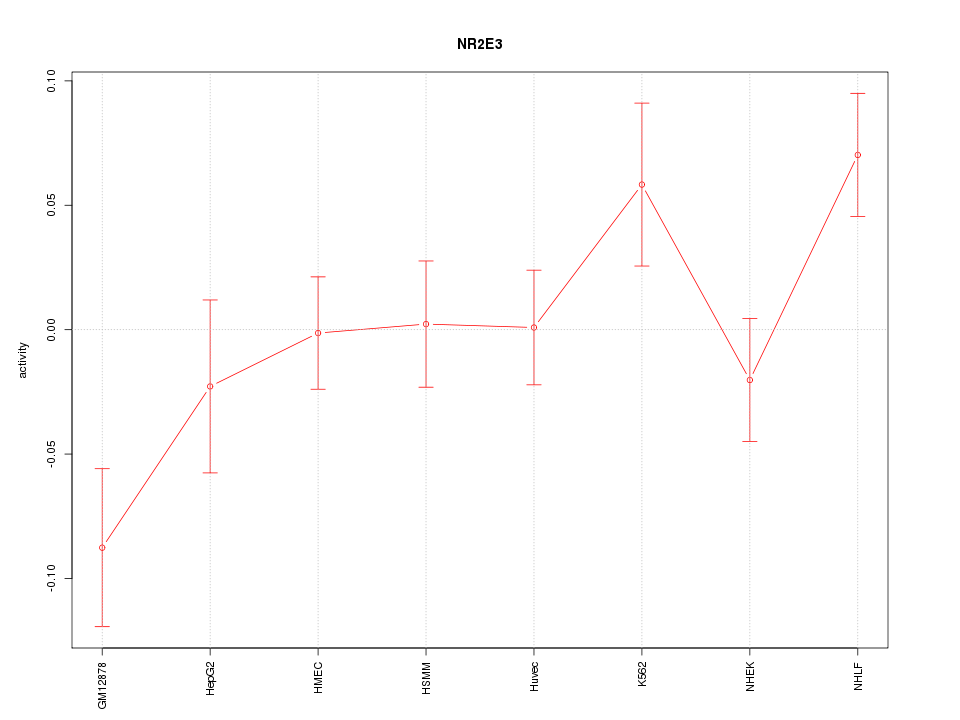
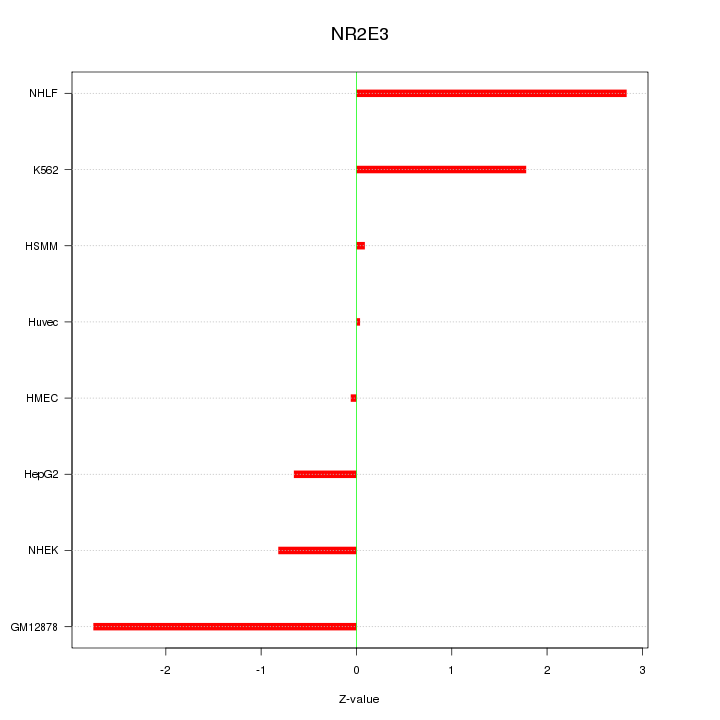
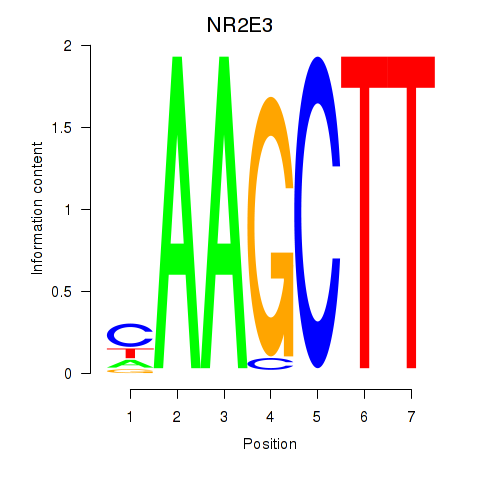
Motif ID: NR2E3
Z-value: 1.579
Transcription factors associated with NR2E3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NR2E3 | ENSG00000031544.10 | NR2E3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.6 | 2.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.4 | 3.2 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.3 | 2.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.3 | 0.8 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 1.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.0 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 1.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.9 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.2 | 0.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.8 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 1.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 2.9 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.5 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 1.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 2.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 2.1 | GO:0003407 | neural retina development(GO:0003407) |
| 0.1 | 0.4 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 1.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 2.1 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.5 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.1 | 0.7 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.9 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.7 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.4 | GO:0001934 | positive regulation of protein phosphorylation(GO:0001934) |
| 0.1 | 0.2 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.1 | 0.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.0 | GO:0033687 | osteoblast proliferation(GO:0033687) negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.2 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 1.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 2.7 | GO:0050931 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.1 | 0.2 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) sensory perception of umami taste(GO:0050917) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.8 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.2 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.1 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.2 | GO:0002331 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.0 | 0.3 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 1.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0051255 | spindle midzone assembly(GO:0051255) meiotic spindle midzone assembly(GO:0051257) |
| 0.0 | 0.8 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0045759 | optic nerve morphogenesis(GO:0021631) regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) negative regulation of action potential(GO:0045759) synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.3 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 1.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.3 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.9 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:0045760 | operant conditioning(GO:0035106) positive regulation of action potential(GO:0045760) |
| 0.0 | 0.4 | GO:0048255 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 1.0 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0031670 | cellular response to nutrient(GO:0031670) |
| 0.0 | 0.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 2.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.8 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.1 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 2.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 1.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) MutLbeta complex(GO:0032390) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 2.0 | GO:1902554 | serine/threonine protein kinase complex(GO:1902554) |
| 0.0 | 0.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 1.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 2.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 1.6 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 1.4 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.3 | 2.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 2.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 1.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 0.6 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.3 | GO:0050816 | beta-2 adrenergic receptor binding(GO:0031698) phosphothreonine binding(GO:0050816) |
| 0.1 | 1.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 3.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.4 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 0.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.3 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 3.1 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 1.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0046934 | obsolete inositol or phosphatidylinositol kinase activity(GO:0004428) phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 1.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.7 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0019894 | kinesin binding(GO:0019894) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.7 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |