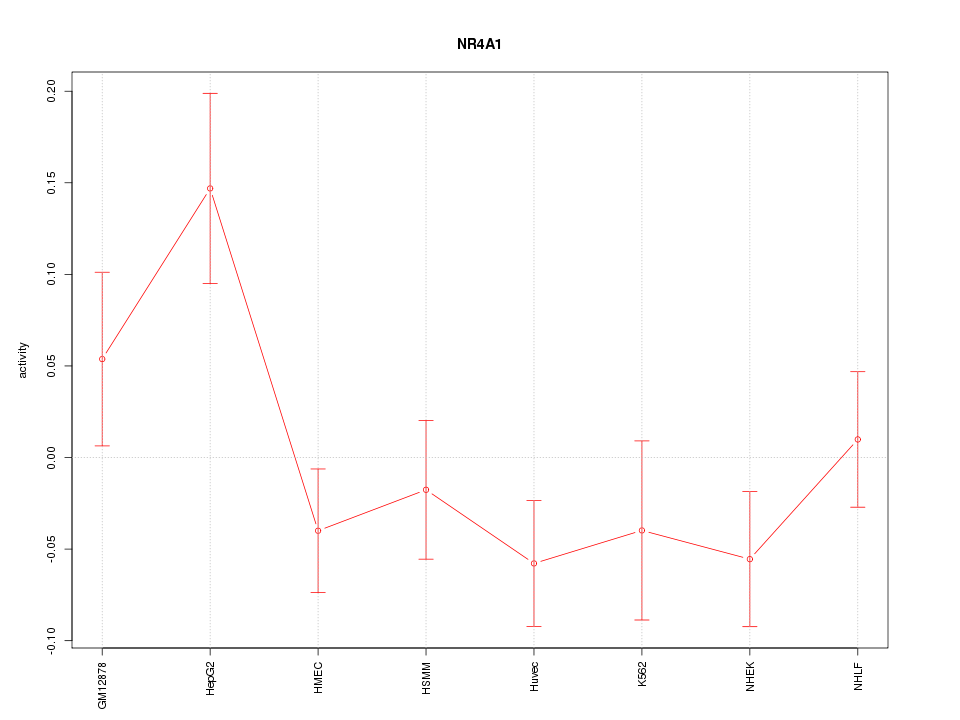
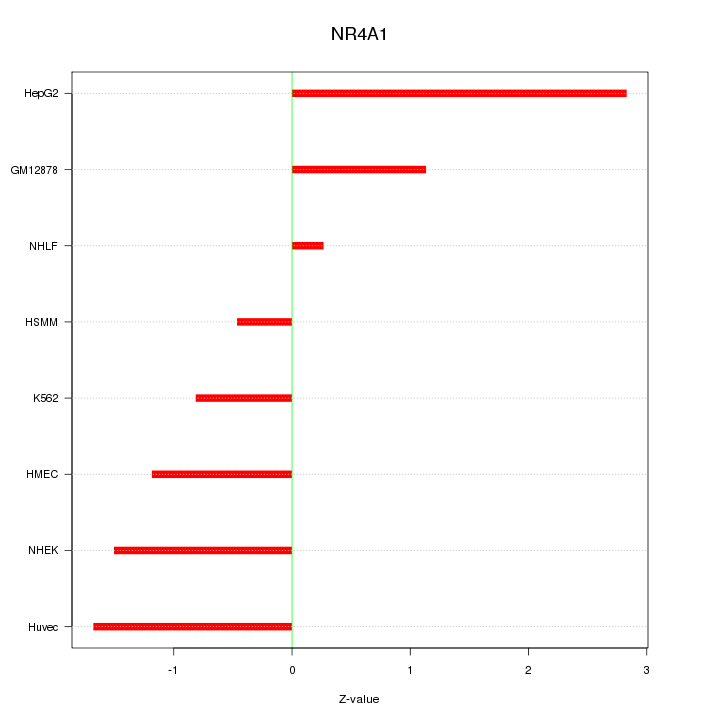
Motif ID: NR4A1
Z-value: 1.446
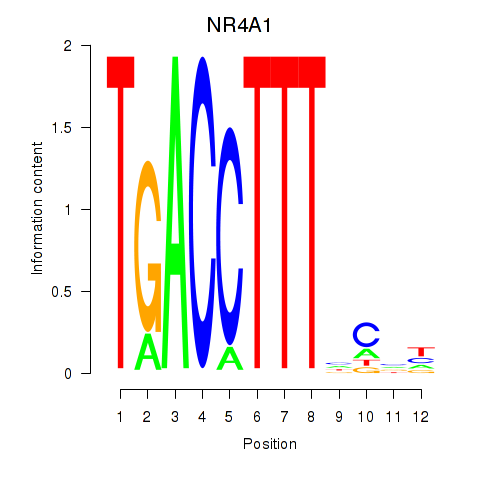
Transcription factors associated with NR4A1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NR4A1 | ENSG00000123358.15 | NR4A1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0010989 | regulation of high-density lipoprotein particle clearance(GO:0010982) negative regulation of low-density lipoprotein particle clearance(GO:0010989) regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.8 | 2.3 | GO:0014063 | regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.6 | 2.6 | GO:0031643 | regulation of Schwann cell differentiation(GO:0014038) positive regulation of Schwann cell differentiation(GO:0014040) positive regulation of myelination(GO:0031643) |
| 0.4 | 2.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 | 1.8 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 1.8 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 3.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.6 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.2 | 1.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.3 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 1.4 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.1 | 0.4 | GO:2001259 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.1 | 0.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.7 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) obsolete negative regulation of diuresis(GO:0003077) positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.4 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 4.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 1.2 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 1.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.8 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.5 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 1.3 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 1.6 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.3 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 1.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 2.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 3.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0043197 | dendritic spine(GO:0043197) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.6 | 2.6 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.5 | 2.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 1.5 | GO:0008907 | integrase activity(GO:0008907) |
| 0.4 | 3.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 2.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 1.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.9 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 1.8 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.5 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 4.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0015141 | thiosulfate transmembrane transporter activity(GO:0015117) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 2.3 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.8 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 2.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0060590 | ATPase regulator activity(GO:0060590) |