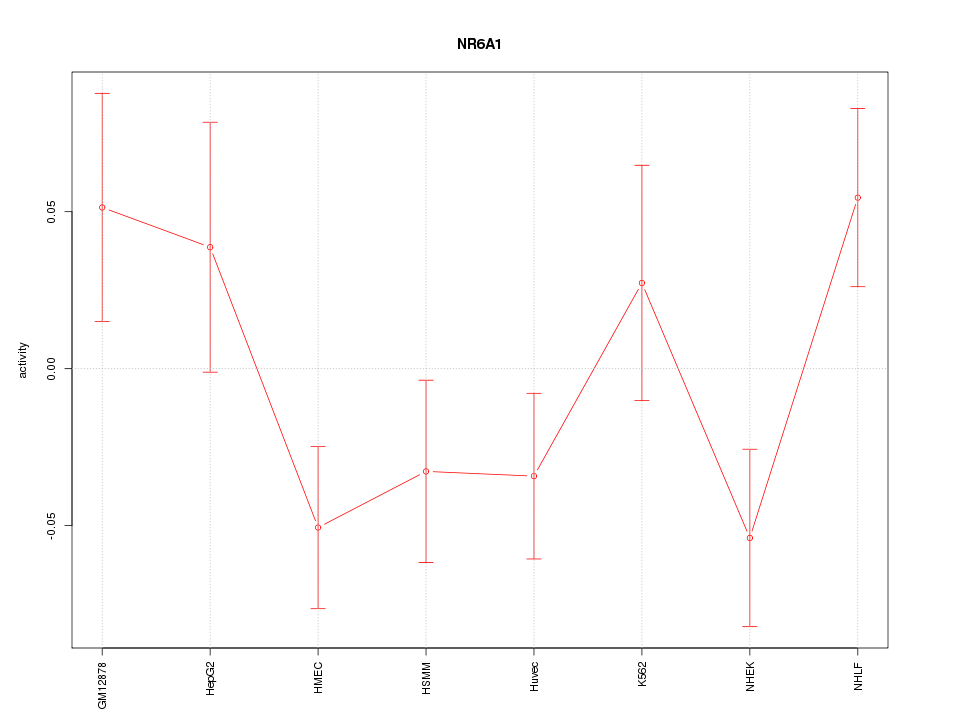
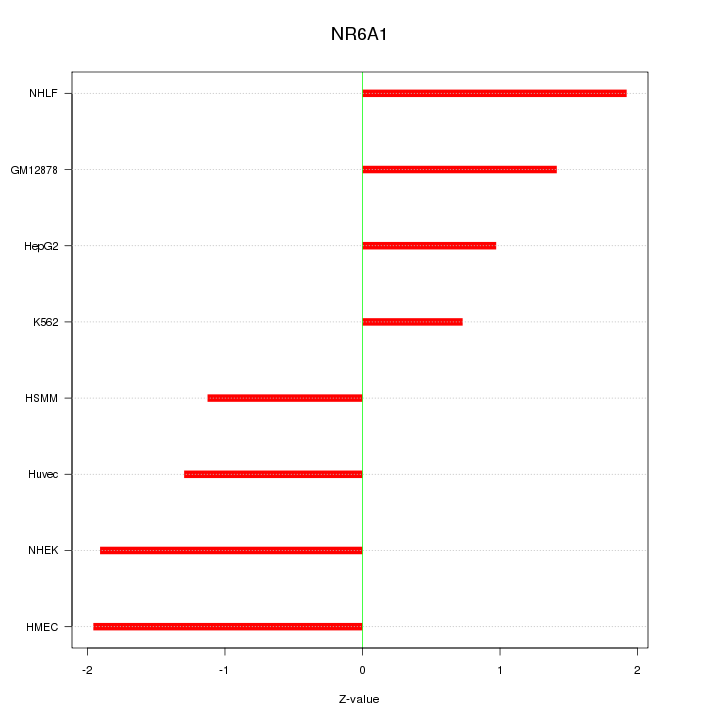
Motif ID: NR6A1
Z-value: 1.483
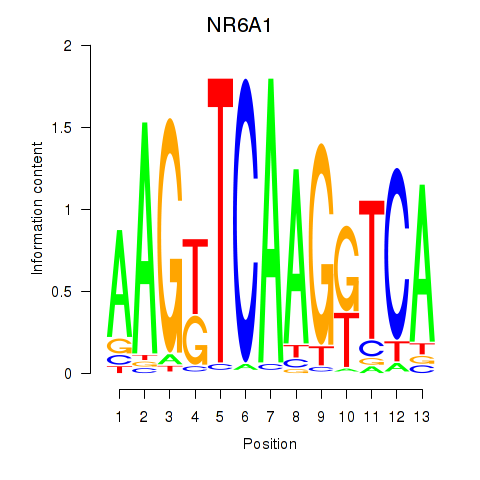
Transcription factors associated with NR6A1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NR6A1 | ENSG00000148200.12 | NR6A1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.3 | 0.9 | GO:0014063 | regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.3 | 1.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.3 | 1.8 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 0.8 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.2 | 0.7 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.2 | 1.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.9 | GO:0034442 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.2 | 0.6 | GO:0060267 | transformation of host cell by virus(GO:0019087) positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 0.8 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.6 | GO:0070527 | integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) platelet aggregation(GO:0070527) |
| 0.1 | 0.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.5 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.5 | GO:0003070 | age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.1 | 1.2 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) |
| 0.1 | 0.7 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 0.3 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 1.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.7 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 0.2 | GO:0002124 | territorial aggressive behavior(GO:0002124) brainstem development(GO:0003360) vocalization behavior(GO:0071625) |
| 0.1 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.6 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.4 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 10.2 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.1 | 1.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.6 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.8 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.7 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.4 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0007285 | primary spermatocyte growth(GO:0007285) |
| 0.0 | 0.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.9 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 1.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0060157 | urea transport(GO:0015840) urinary bladder development(GO:0060157) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:2001259 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.0 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.4 | GO:0044349 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.0 | 0.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.9 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.6 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.9 | GO:0006094 | gluconeogenesis(GO:0006094) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0036125 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 0.3 | 1.8 | GO:0019815 | immunoglobulin complex(GO:0019814) B cell receptor complex(GO:0019815) |
| 0.2 | 2.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.5 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.0 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.0 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.0 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0004917 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) interleukin-2 binding(GO:0019976) |
| 0.3 | 0.8 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.3 | 0.8 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.2 | 1.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.7 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.2 | 0.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.3 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 1.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.4 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.8 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.1 | 0.2 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.1 | 0.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.0 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |