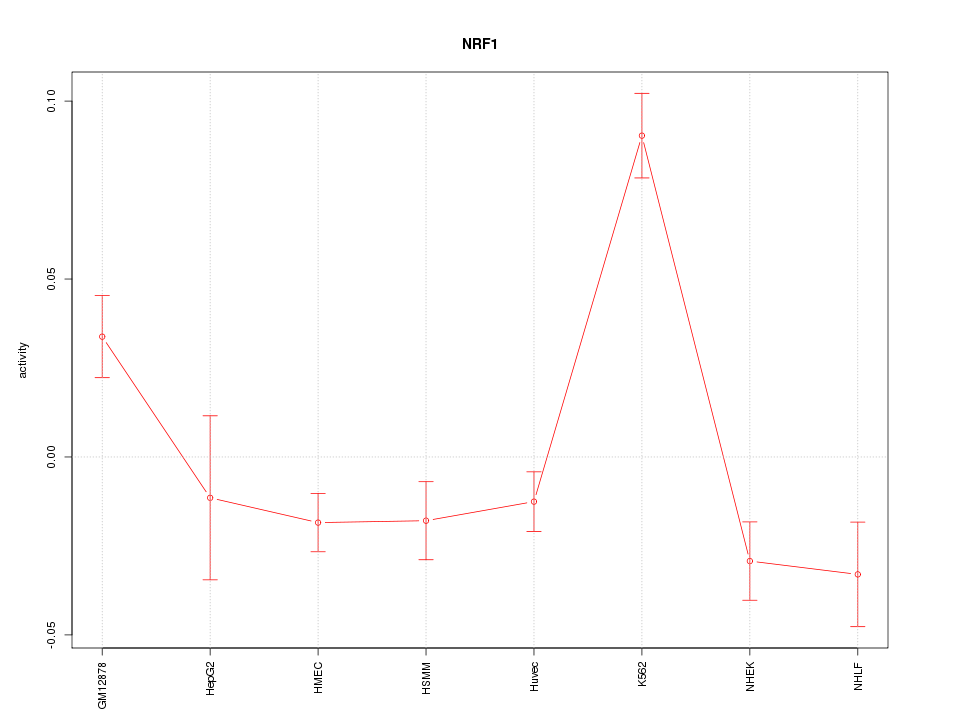
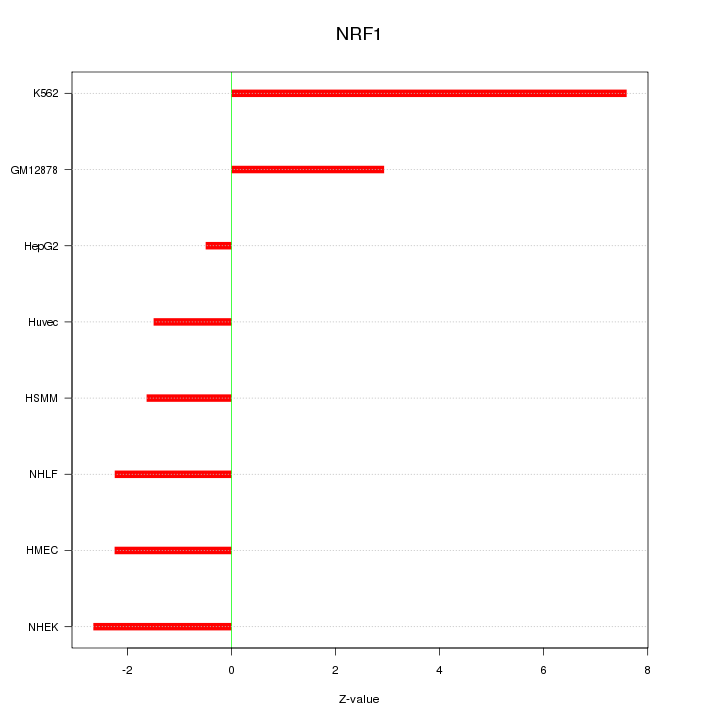
Motif ID: NRF1
Z-value: 3.328
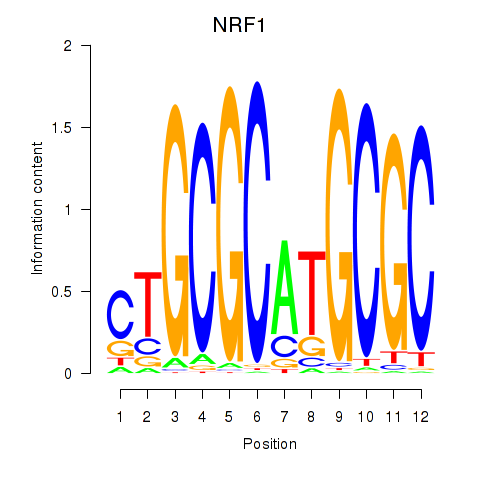
Transcription factors associated with NRF1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NRF1 | ENSG00000106459.10 | NRF1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.9 | 2.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.8 | 2.3 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.7 | 5.5 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.7 | 8.3 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.7 | 2.1 | GO:0051255 | spindle midzone assembly(GO:0051255) meiotic spindle midzone assembly(GO:0051257) |
| 0.7 | 2.6 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.6 | 3.0 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.6 | 2.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.5 | 1.1 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.5 | 2.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.5 | 2.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.5 | 1.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.5 | 1.9 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.5 | 4.3 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.5 | 1.4 | GO:0010041 | response to iron(III) ion(GO:0010041) positive regulation of histone phosphorylation(GO:0033129) response to DDT(GO:0046680) regulation of chromosome condensation(GO:0060623) cellular response to iron ion(GO:0071281) cellular response to iron(III) ion(GO:0071283) |
| 0.5 | 1.8 | GO:0007262 | STAT protein import into nucleus(GO:0007262) regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.5 | 1.8 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) intra-S DNA damage checkpoint(GO:0031573) negative regulation of meiotic nuclear division(GO:0045835) regulation of meiosis I(GO:0060631) |
| 0.4 | 1.3 | GO:1990170 | detoxification of cadmium ion(GO:0071585) stress response to cadmium ion(GO:1990170) |
| 0.4 | 1.3 | GO:0014889 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) muscle atrophy(GO:0014889) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.4 | 1.3 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) protein hexamerization(GO:0034214) |
| 0.4 | 1.3 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.4 | 2.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.4 | 1.2 | GO:1904742 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) regulation of telomeric DNA binding(GO:1904742) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.4 | 2.0 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.4 | 3.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.4 | 1.6 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.4 | 7.8 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.4 | 2.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 1.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 1.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.4 | 1.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.4 | 6.4 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.3 | 3.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 5.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.3 | 2.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.3 | 2.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.3 | 1.3 | GO:0016233 | telomere capping(GO:0016233) |
| 0.3 | 1.0 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.3 | 1.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 1.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 0.3 | 2.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 1.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.3 | 1.8 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.3 | 0.6 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.3 | 1.2 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.3 | 0.8 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.3 | 2.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 2.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.3 | 1.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 0.8 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.3 | 0.8 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.3 | 1.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 6.0 | GO:0031055 | DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 4.7 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.3 | 1.6 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.3 | 0.8 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.3 | 0.8 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.3 | 6.1 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 1.5 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.3 | 0.3 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.3 | 0.8 | GO:0031297 | replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.2 | 1.7 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.2 | 0.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.2 | 2.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 1.4 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.2 | 1.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 1.6 | GO:0015853 | adenine transport(GO:0015853) |
| 0.2 | 1.9 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.2 | 0.7 | GO:0071104 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.2 | 1.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 2.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 1.6 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.2 | 2.5 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.2 | 0.7 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.2 | 1.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 0.9 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 0.9 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.2 | 0.9 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.2 | 0.9 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 0.8 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 2.3 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.2 | 2.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 1.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 1.4 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.2 | 0.6 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 0.8 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.2 | 2.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 1.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.2 | 1.4 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) |
| 0.2 | 1.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.2 | 13.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 0.8 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 2.8 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.2 | 0.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 0.7 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.2 | 0.6 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 0.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 1.4 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.2 | 1.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 1.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 3.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 5.4 | GO:0050684 | regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.2 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 2.7 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.2 | 1.3 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.2 | 1.3 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 0.5 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.2 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 0.7 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.2 | 0.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 2.0 | GO:0044349 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.2 | 0.8 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.2 | 0.3 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.1 | 1.2 | GO:0039703 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 1.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.4 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 1.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 1.6 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.1 | 0.9 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 2.3 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 | 0.9 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 2.0 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 1.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 1.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.7 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.4 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 6.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.4 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) hair follicle placode formation(GO:0060789) fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.1 | 1.1 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.1 | 0.4 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 0.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 8.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.1 | 0.5 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 1.0 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 3.6 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.6 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 1.0 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 3.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.4 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.1 | 0.4 | GO:0034442 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.1 | 0.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 0.2 | GO:0042775 | ATP synthesis coupled electron transport(GO:0042773) mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 1.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.5 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 0.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 1.0 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.4 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.9 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.1 | 1.3 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 3.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.2 | GO:0031062 | positive regulation of histone methylation(GO:0031062) positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.1 | 0.4 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 1.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.4 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 2.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.1 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.1 | 0.3 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 2.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.6 | GO:0031958 | corticosteroid receptor signaling pathway(GO:0031958) |
| 0.1 | 0.5 | GO:0003070 | age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.1 | 4.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0051383 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.1 | 0.7 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.6 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.6 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.7 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 1.8 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 2.9 | GO:0034470 | ncRNA processing(GO:0034470) |
| 0.1 | 0.5 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.2 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.1 | 2.2 | GO:0006490 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 2.0 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.1 | 0.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.4 | GO:0060676 | ureteric bud formation(GO:0060676) mesonephric tubule formation(GO:0072172) |
| 0.1 | 2.2 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 2.6 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 1.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.4 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.2 | GO:0070781 | response to biotin(GO:0070781) |
| 0.1 | 1.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 3.5 | GO:0007051 | spindle organization(GO:0007051) |
| 0.1 | 2.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 2.5 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.3 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 1.3 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 3.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.2 | GO:0021819 | cerebral cortex radial glia guided migration(GO:0021801) layer formation in cerebral cortex(GO:0021819) telencephalon glial cell migration(GO:0022030) |
| 0.1 | 7.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:0009451 | RNA modification(GO:0009451) |
| 0.1 | 0.9 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.3 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 | 1.0 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.1 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.3 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.7 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.1 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.5 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.3 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.3 | GO:0032527 | retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.1 | 1.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.4 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 1.1 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 0.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.1 | 0.7 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.4 | GO:0016571 | histone methylation(GO:0016571) |
| 0.0 | 1.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 3.1 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.1 | GO:0072148 | epithelial cell fate commitment(GO:0072148) |
| 0.0 | 1.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.2 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.4 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.2 | GO:0045350 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 1.4 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.0 | 13.1 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.3 | GO:0051223 | regulation of protein transport(GO:0051223) |
| 0.0 | 1.2 | GO:0019674 | NAD metabolic process(GO:0019674) |
| 0.0 | 2.2 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.5 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 1.2 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 2.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.7 | GO:0070646 | protein modification by small protein removal(GO:0070646) |
| 0.0 | 0.9 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.2 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.3 | GO:0031670 | cellular response to nutrient(GO:0031670) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.3 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.6 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.3 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.6 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.7 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 0.4 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0071679 | facial nucleus development(GO:0021754) cell proliferation in midbrain(GO:0033278) commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.5 | GO:0071333 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.2 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 2.6 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.7 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.9 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.6 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.7 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.9 | GO:0051320 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.0 | 0.5 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.7 | GO:0009067 | aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.0 | 1.4 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.3 | GO:0051329 | mitotic interphase(GO:0051329) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.0 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.2 | GO:0045060 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.4 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.2 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) DNA methylation or demethylation(GO:0044728) |
| 0.0 | 0.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.6 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.1 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.5 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.4 | GO:0001756 | somitogenesis(GO:0001756) somite development(GO:0061053) |
| 0.0 | 0.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.5 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.0 | 1.3 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.3 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.5 | GO:1903046 | meiotic cell cycle(GO:0051321) meiotic cell cycle process(GO:1903046) |
| 0.0 | 0.1 | GO:0046632 | alpha-beta T cell differentiation(GO:0046632) |
| 0.0 | 0.8 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.4 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.2 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.2 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.8 | GO:0034339 | obsolete regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor(GO:0034339) |
| 0.0 | 0.3 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.3 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.0 | 0.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0048167 | regulation of synaptic plasticity(GO:0048167) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.3 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 0.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0030593 | neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.0 | 0.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 2.7 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 1.5 | 8.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.8 | 2.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.6 | 1.8 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.6 | 2.3 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.5 | 2.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.5 | 3.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.4 | 1.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.4 | 2.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.4 | 3.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.4 | 1.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.4 | 1.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 2.8 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.3 | 2.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 1.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 4.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.3 | 2.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 1.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 0.9 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.3 | 0.9 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 1.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.3 | 2.5 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.3 | 2.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 1.4 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.3 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 6.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 2.6 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 13.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 2.0 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.3 | 0.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 3.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 0.7 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.2 | 3.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 3.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 1.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 1.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 2.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 0.8 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.2 | 1.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.0 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.2 | 3.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.2 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.8 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.2 | 1.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.2 | 1.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 1.9 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 14.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 1.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 0.5 | GO:0036452 | ESCRT III complex(GO:0000815) ESCRT complex(GO:0036452) |
| 0.2 | 0.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 0.6 | GO:0001940 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.1 | 0.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) U4/U6 snRNP(GO:0071001) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.1 | 1.2 | GO:0005719 | euchromatin(GO:0000791) nuclear euchromatin(GO:0005719) |
| 0.1 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 2.0 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.1 | 1.5 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 1.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 6.3 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 4.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 1.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 1.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 2.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 1.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 5.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 9.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.8 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 2.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 4.3 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.2 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.1 | 1.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 4.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 2.4 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.4 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 4.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.5 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.1 | 0.5 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 2.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 3.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.3 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 3.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 6.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.5 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 3.5 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 28.9 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 2.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 4.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.6 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 0.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.8 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.3 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) axoneme part(GO:0044447) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.9 | 2.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.8 | 6.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.8 | 2.3 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.7 | 8.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.6 | 1.8 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.6 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.6 | 2.3 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.6 | 1.7 | GO:0070260 | 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.6 | 0.6 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.5 | 1.5 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.5 | 1.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.5 | 1.4 | GO:0015141 | thiosulfate transmembrane transporter activity(GO:0015117) succinate transmembrane transporter activity(GO:0015141) |
| 0.4 | 1.3 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.4 | 0.9 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.4 | 2.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.4 | 1.7 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.4 | 2.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.4 | 1.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.4 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.4 | 1.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.4 | 1.9 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 1.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.4 | 1.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 3.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.3 | 1.0 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.3 | 0.9 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 1.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 1.2 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.3 | 4.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 1.9 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.3 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 1.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 0.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 0.5 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.3 | 1.4 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 2.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 1.0 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.3 | 0.8 | GO:0043426 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) MRF binding(GO:0043426) |
| 0.3 | 0.8 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.3 | 4.6 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.2 | 1.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 2.4 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.2 | 0.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 11.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 0.5 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 0.9 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.2 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 1.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 2.7 | GO:0004526 | ribonuclease P activity(GO:0004526) tRNA-specific ribonuclease activity(GO:0004549) |
| 0.2 | 0.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 1.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.2 | 0.9 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 0.9 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.2 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.9 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.2 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 0.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 0.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 5.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.2 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 4.7 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.2 | 1.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.2 | 0.5 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 1.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 1.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 2.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.6 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 2.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.5 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.1 | GO:0030611 | arsenate reductase activity(GO:0030611) |
| 0.1 | 2.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.9 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 1.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.7 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.5 | GO:0008148 | obsolete negative transcription elongation factor activity(GO:0008148) |
| 0.1 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.5 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.7 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 5.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 4.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 2.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.9 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 4.7 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.9 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.3 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.1 | 0.5 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 8.8 | GO:0016566 | obsolete specific transcriptional repressor activity(GO:0016566) |
| 0.1 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.1 | 0.8 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 0.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 2.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.8 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.8 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 1.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.0 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.4 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.1 | 0.9 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.0 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 0.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.7 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 3.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.1 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 0.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 0.3 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) deoxyribonucleotide binding(GO:0032552) |
| 0.1 | 2.3 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.2 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.3 | GO:0004428 | obsolete inositol or phosphatidylinositol kinase activity(GO:0004428) |
| 0.1 | 0.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 3.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.6 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 4.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.4 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 1.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.9 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 1.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 5.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 3.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.8 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.2 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.9 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 4.1 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 3.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.1 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 2.7 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.4 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.3 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.3 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 3.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 2.5 | GO:0010843 | obsolete promoter binding(GO:0010843) |
| 0.0 | 0.2 | GO:0016741 | transferase activity, transferring one-carbon groups(GO:0016741) |
| 0.0 | 3.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 4.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 1.5 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 4.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.0 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.7 | GO:0019207 | kinase regulator activity(GO:0019207) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 1.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.2 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.3 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.7 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 3.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.1 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 17.9 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.1 | 2.2 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.1 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.6 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.3 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.1 | 0.5 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.3 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.1 | 0.7 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.7 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.1 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |