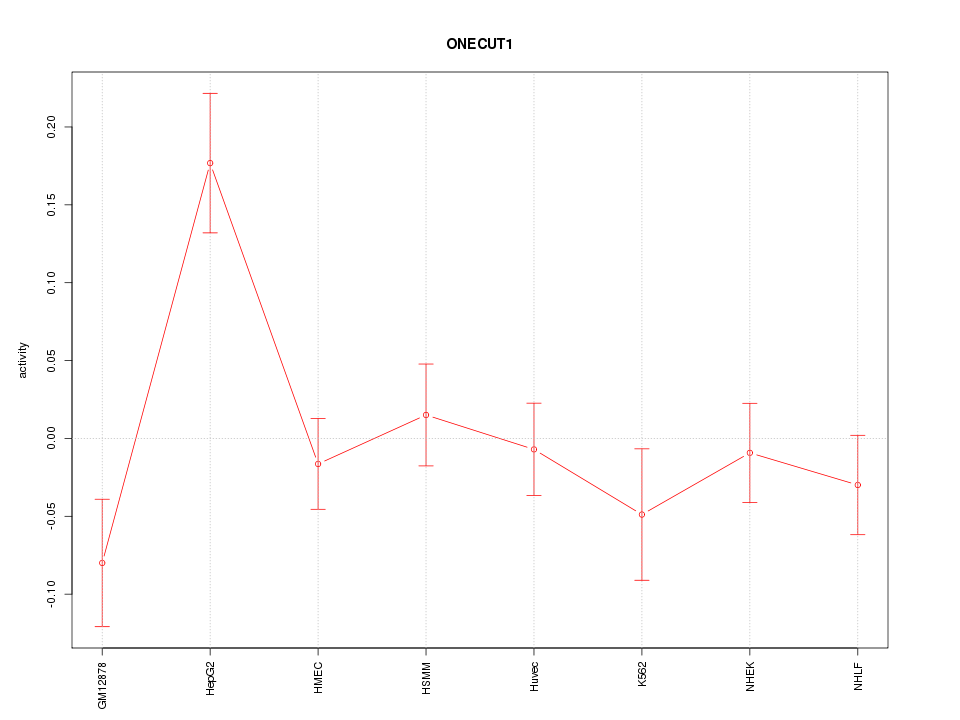
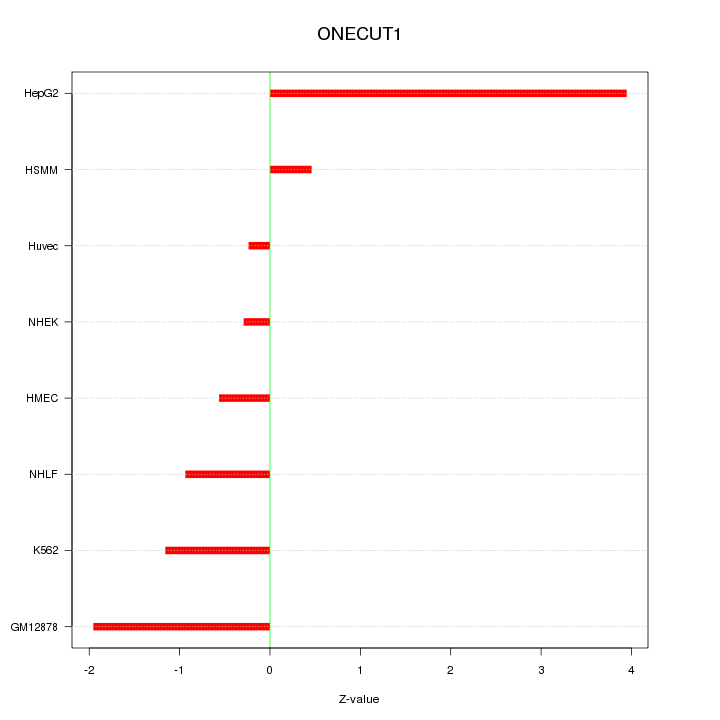
Motif ID: ONECUT1
Z-value: 1.669
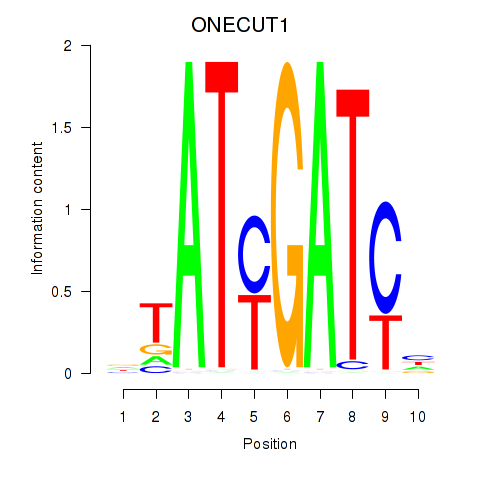
Transcription factors associated with ONECUT1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ONECUT1 | ENSG00000169856.7 | ONECUT1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) positive regulation of phospholipid catabolic process(GO:0060697) |
| 1.6 | 15.6 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.8 | 3.1 | GO:0001705 | ectoderm formation(GO:0001705) central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.7 | 2.2 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.7 | 2.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.6 | 1.9 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.5 | 4.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.5 | 1.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.4 | 1.3 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.4 | 5.8 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 1.6 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.4 | 2.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 1.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.4 | 2.4 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 | 1.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.2 | 13.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 1.2 | GO:0015827 | tryptophan transport(GO:0015827) leucine import(GO:0060356) |
| 0.2 | 0.8 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.2 | 2.6 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.2 | 0.8 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.4 | GO:2000858 | renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) positive regulation of NAD(P)H oxidase activity(GO:0033864) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.1 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 0.7 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.1 | 1.5 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.9 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.8 | GO:0036503 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 0.5 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.0 | 0.4 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.3 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 1.2 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.9 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.2 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 1.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.4 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 2.1 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 0.4 | GO:0008038 | neuron recognition(GO:0008038) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 10.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.5 | 2.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 3.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 2.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 1.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 9.0 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 2.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 14.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 6.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 14.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.4 | 5.8 | GO:0019862 | IgA binding(GO:0019862) |
| 1.2 | 4.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.5 | 11.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 2.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 2.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 1.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 2.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.2 | 1.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 2.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 3.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 1.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 3.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.3 | GO:0004608 | phosphatidylethanolamine N-methyltransferase activity(GO:0004608) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.5 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 5.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) fatty acid ligase activity(GO:0015645) |
| 0.0 | 0.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.7 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.2 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 2.7 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 1.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.7 | GO:0004386 | helicase activity(GO:0004386) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.0 | 1.5 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |