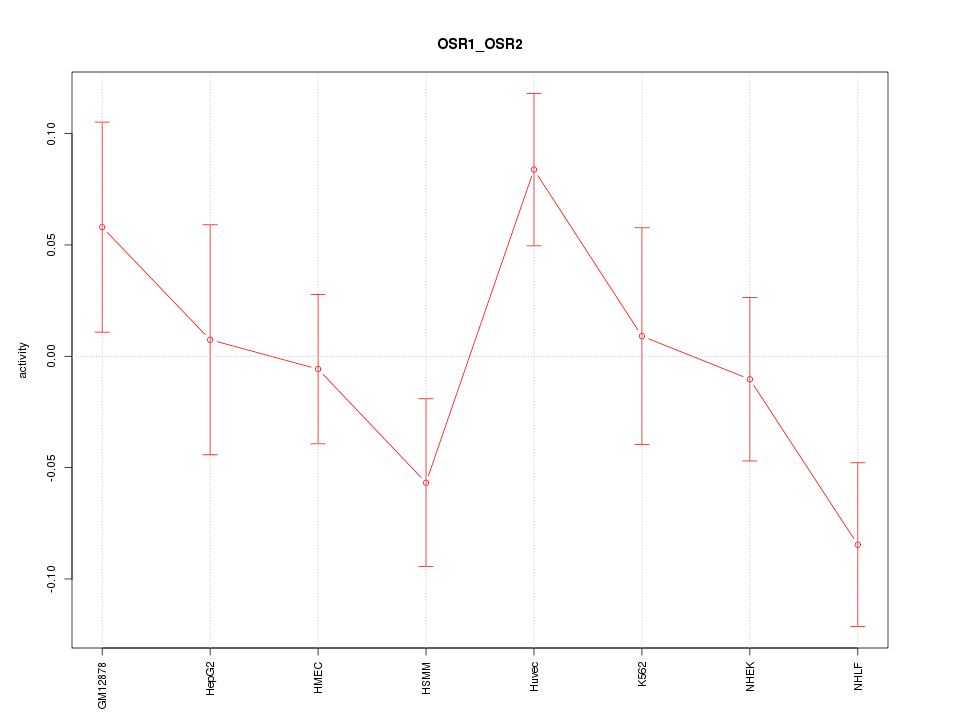
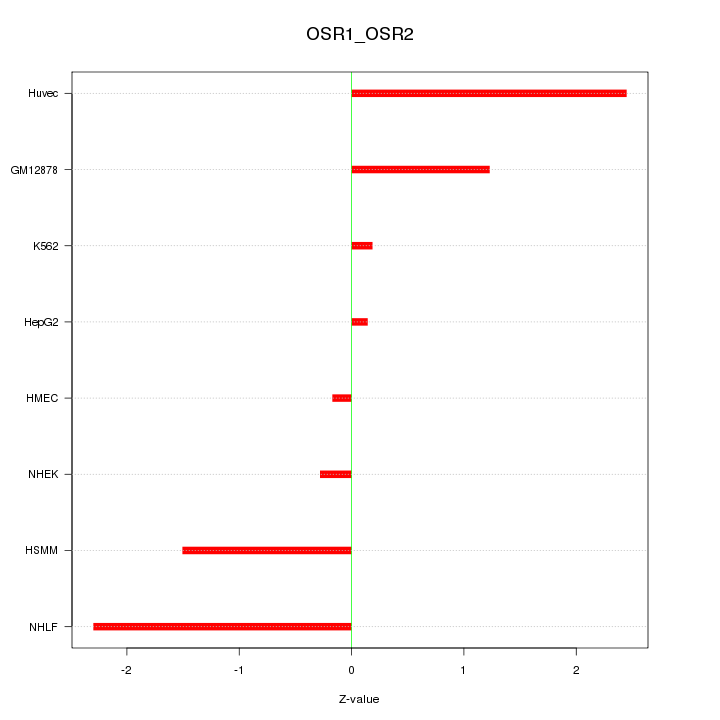
Motif ID: OSR1_OSR2
Z-value: 1.379
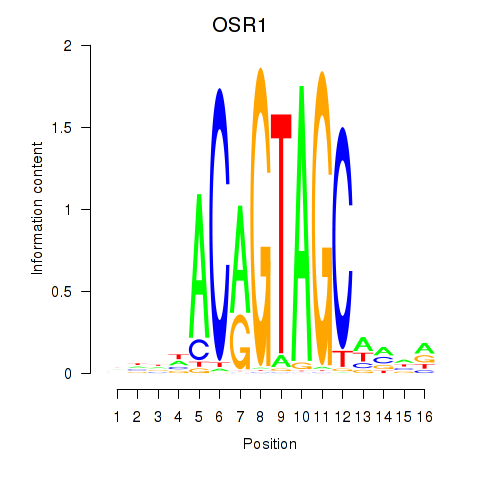
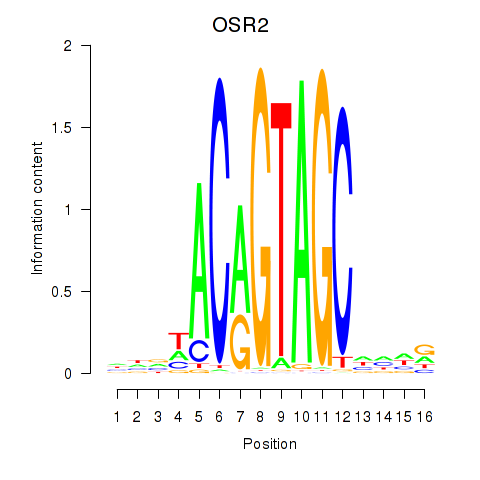
Transcription factors associated with OSR1_OSR2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| OSR1 | ENSG00000143867.5 | OSR1 |
| OSR2 | ENSG00000164920.5 | OSR2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.6 | 1.9 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.4 | 1.1 | GO:0010041 | response to iron(III) ion(GO:0010041) positive regulation of histone phosphorylation(GO:0033129) response to DDT(GO:0046680) regulation of chromosome condensation(GO:0060623) cellular response to iron ion(GO:0071281) cellular response to iron(III) ion(GO:0071283) |
| 0.1 | 0.4 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.1 | 0.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.5 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.1 | 0.4 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.6 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.3 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 7.2 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.3 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 1.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.4 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.7 | 2.1 | GO:0070283 | lipoate-protein ligase activity(GO:0016979) lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.5 | 1.9 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.4 | 1.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 0.5 | GO:0016731 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.3 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 3.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |