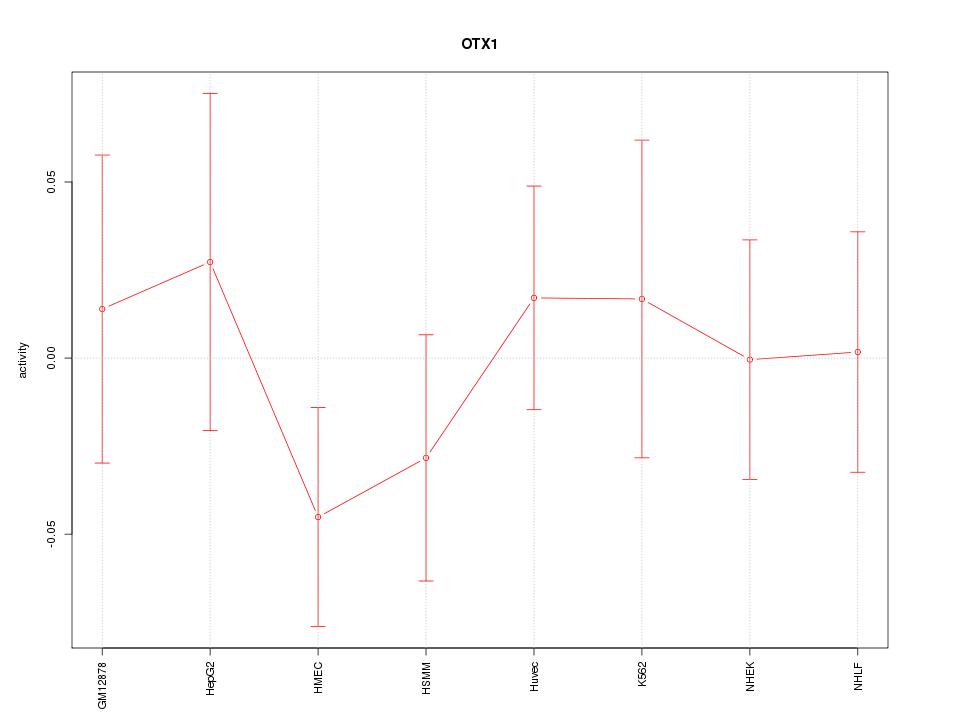
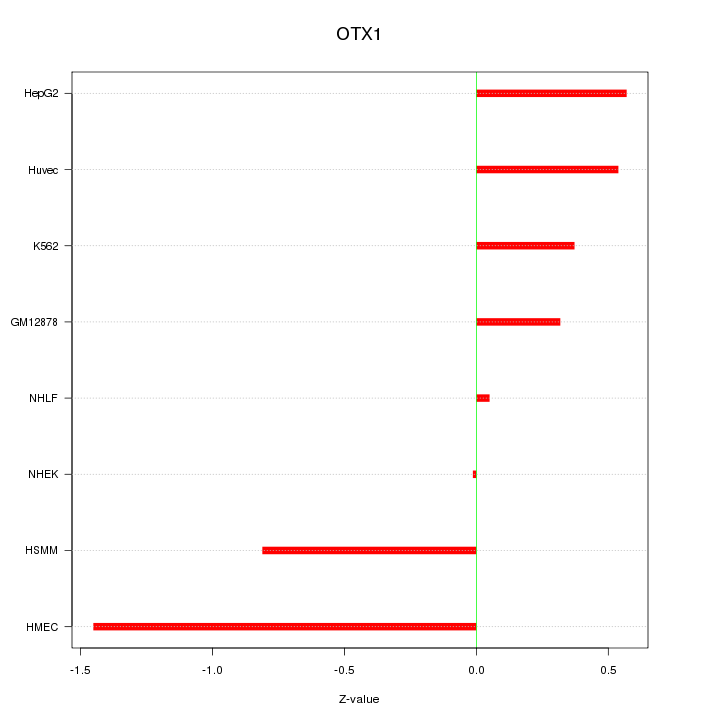
Motif ID: OTX1
Z-value: 0.673
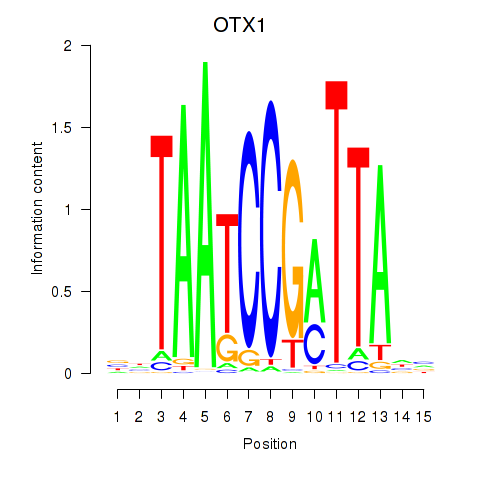
Transcription factors associated with OTX1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| OTX1 | ENSG00000115507.5 | OTX1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.2 | 0.5 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.1 | 0.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.4 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) obsolete negative regulation of diuresis(GO:0003077) positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0031034 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.3 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.2 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.0 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.0 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |