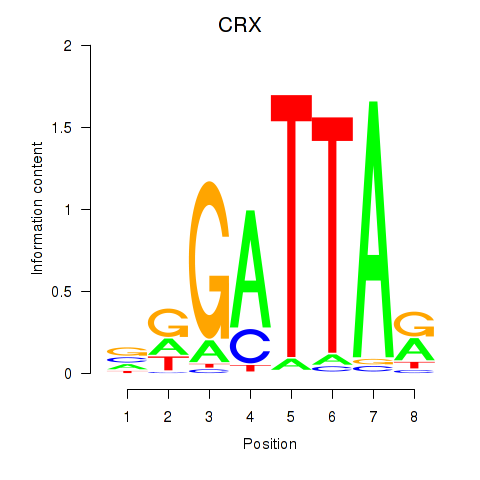
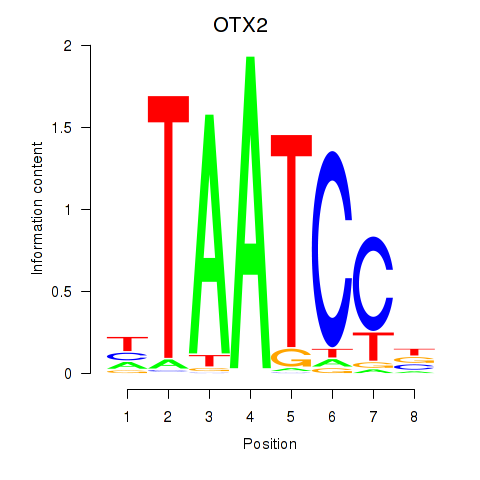
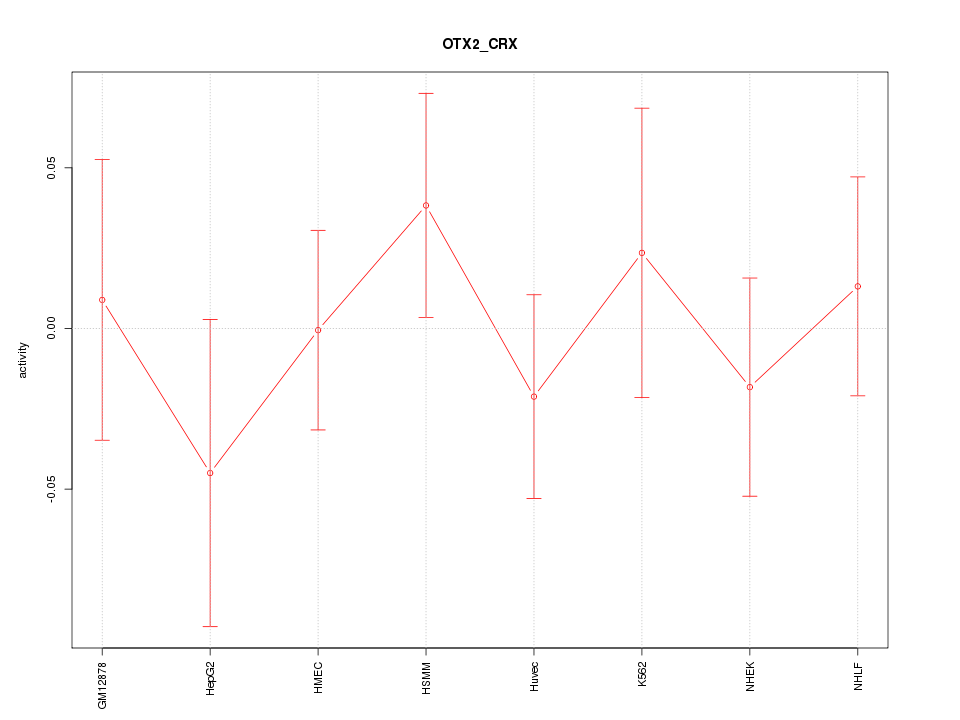
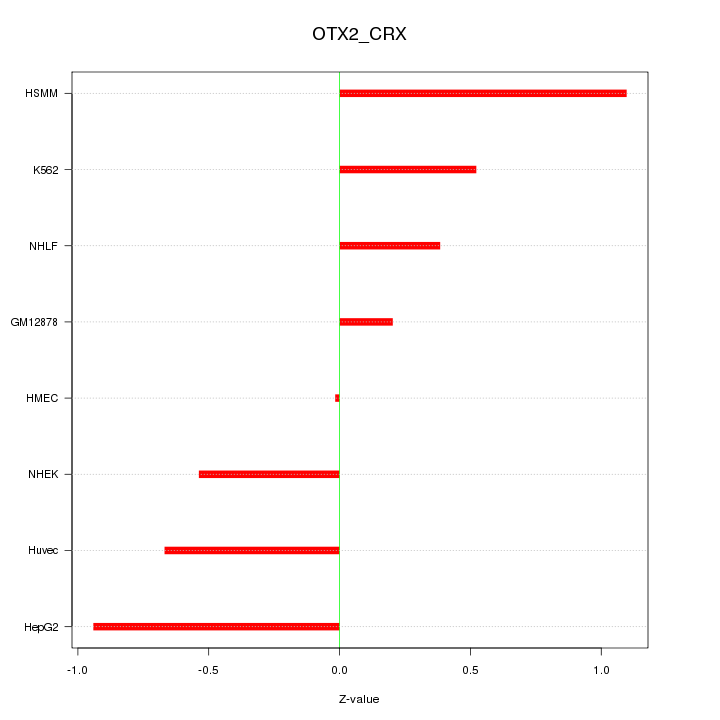
| 0.7 |
2.8 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.1 |
1.3 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
1.0 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 |
0.3 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.0 |
0.4 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.0 |
0.3 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.1 |
GO:0006591 |
ornithine metabolic process(GO:0006591) |
| 0.0 |
0.2 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 |
0.1 |
GO:2000105 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.0 |
0.1 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.0 |
0.2 |
GO:0072205 |
collecting duct development(GO:0072044) metanephric tubule development(GO:0072170) metanephric collecting duct development(GO:0072205) metanephric epithelium development(GO:0072207) metanephric nephron tubule development(GO:0072234) metanephric nephron epithelium development(GO:0072243) |
| 0.0 |
0.1 |
GO:0071380 |
cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 |
0.1 |
GO:0009296 |
obsolete flagellum assembly(GO:0009296) |
| 0.0 |
0.2 |
GO:0015722 |
canalicular bile acid transport(GO:0015722) |
| 0.0 |
0.2 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.4 |
GO:0000052 |
citrulline metabolic process(GO:0000052) arginine catabolic process(GO:0006527) |
| 0.0 |
0.1 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.1 |
GO:0007207 |
phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 |
0.1 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.3 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.1 |
GO:0090267 |
positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 |
0.1 |
GO:0030917 |
midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 |
0.2 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.4 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.0 |
0.1 |
GO:0032528 |
microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 |
0.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.1 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.1 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 |
0.1 |
GO:0019934 |
cGMP-mediated signaling(GO:0019934) regulation of phagocytosis, engulfment(GO:0060099) positive regulation of phagocytosis, engulfment(GO:0060100) regulation of membrane invagination(GO:1905153) positive regulation of membrane invagination(GO:1905155) |
| 0.0 |
1.0 |
GO:0032387 |
negative regulation of intracellular transport(GO:0032387) |
| 0.0 |
0.2 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |