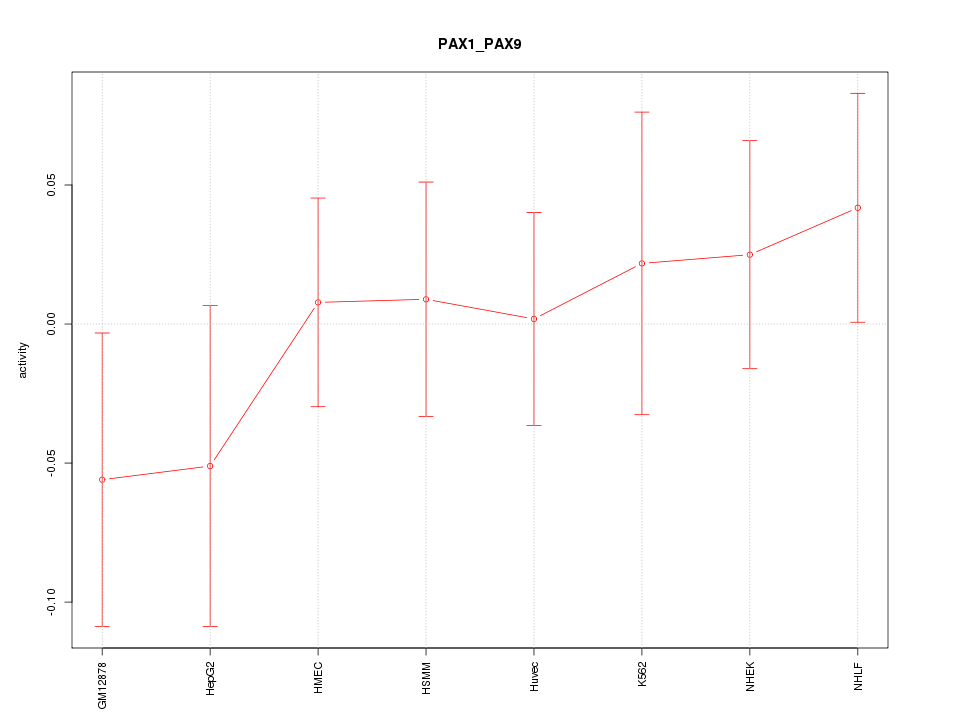
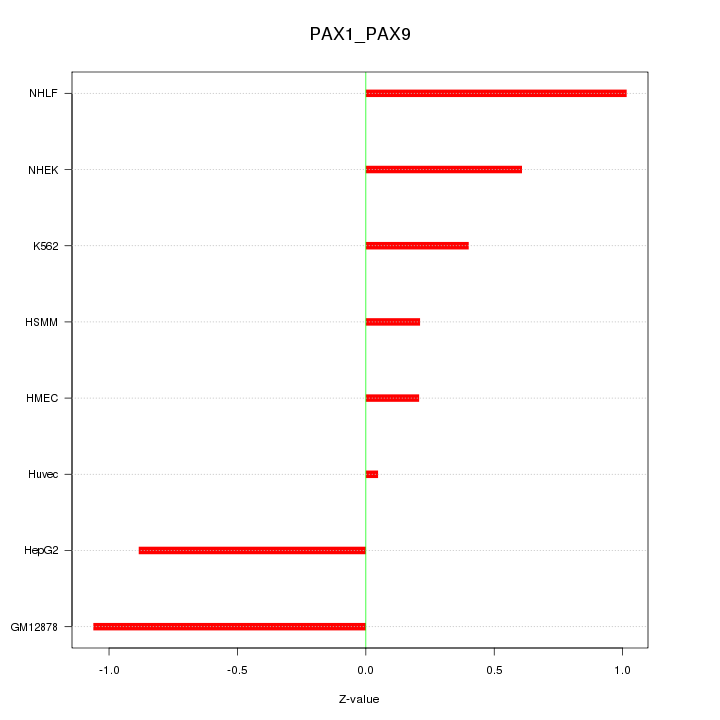
Motif ID: PAX1_PAX9
Z-value: 0.667
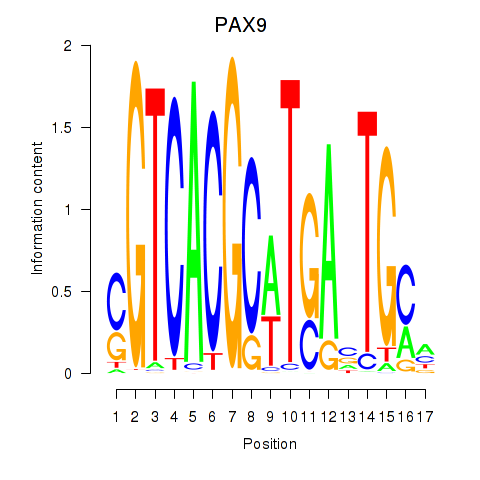
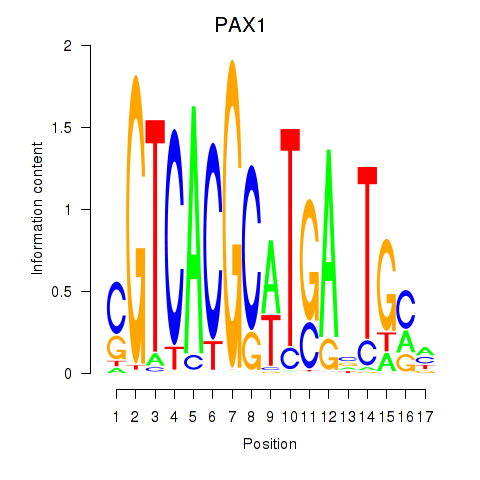
Transcription factors associated with PAX1_PAX9:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PAX1 | ENSG00000125813.9 | PAX1 |
| PAX9 | ENSG00000198807.8 | PAX9 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.3 | 0.8 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.3 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.4 | GO:0010701 | positive regulation of chronic inflammatory response(GO:0002678) positive regulation of norepinephrine secretion(GO:0010701) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) response to muscle activity(GO:0014850) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 1.4 | GO:0043542 | endothelial cell migration(GO:0043542) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |