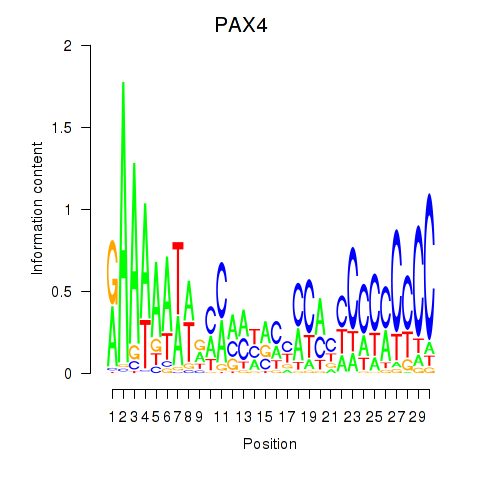
Motif ID: PAX4
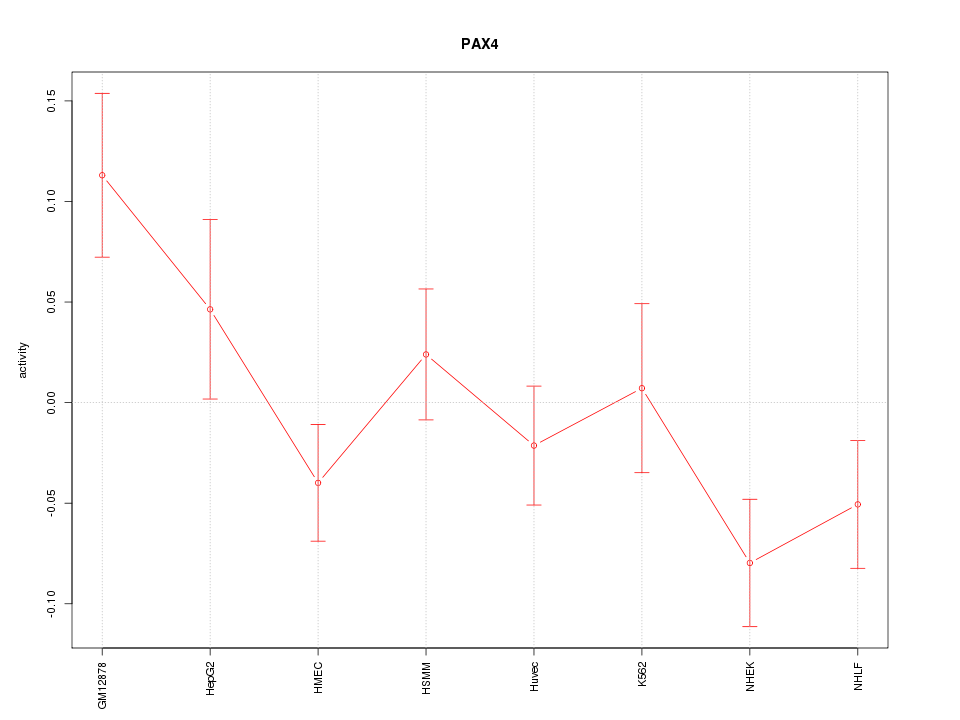
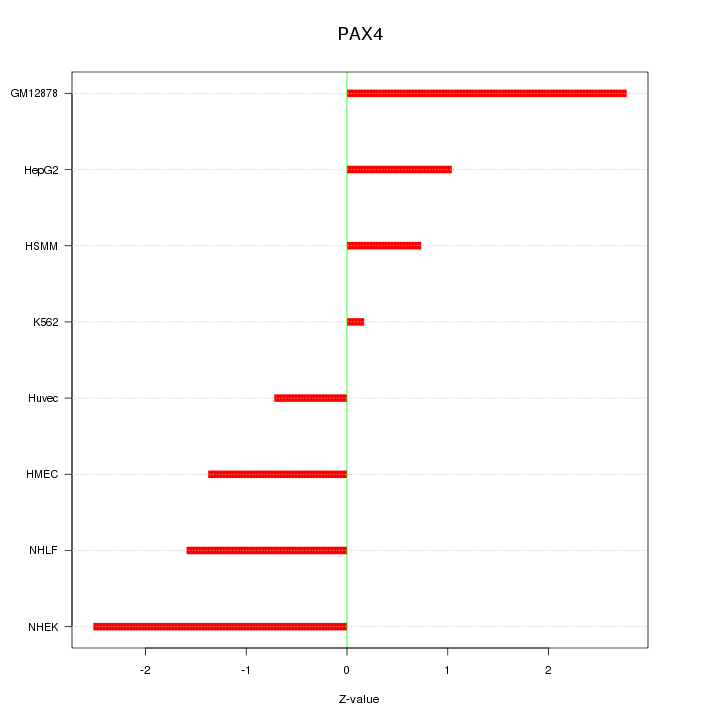
Z-value: 1.606
Transcription factors associated with PAX4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PAX4 | ENSG00000106331.10 | PAX4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.4 | 9.4 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.3 | 1.0 | GO:0046671 | regulation of nitrogen utilization(GO:0006808) positive regulation of neuron maturation(GO:0014042) nitrogen utilization(GO:0019740) cochlear nucleus development(GO:0021747) negative regulation of cellular pH reduction(GO:0032848) glial cell apoptotic process(GO:0034349) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) positive regulation of cell maturation(GO:1903431) |
| 0.3 | 0.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.7 | GO:0034135 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) antibacterial humoral response(GO:0019731) regulation of toll-like receptor 2 signaling pathway(GO:0034135) activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 2.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 0.7 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) negative regulation of meiotic nuclear division(GO:0045835) regulation of meiosis I(GO:0060631) |
| 0.2 | 0.7 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.2 | 0.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 0.7 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.7 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.5 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.5 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 2.8 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 2.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 2.9 | GO:0051923 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) sulfation(GO:0051923) |
| 0.1 | 1.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.4 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.1 | 5.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.8 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.1 | 0.7 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.1 | 1.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 1.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) skeletal muscle organ development(GO:0060538) |
| 0.1 | 0.5 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 1.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.1 | GO:0032650 | bleb assembly(GO:0032060) regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.1 | 1.0 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.1 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0033240 | acetylcholine metabolic process(GO:0008291) positive regulation of cellular amine metabolic process(GO:0033240) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.5 | GO:0098534 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.9 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.2 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.5 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.4 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 0.3 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.0 | 0.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0099515 | nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.4 | GO:0031055 | DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.5 | GO:0009994 | oocyte differentiation(GO:0009994) oocyte development(GO:0048599) |
| 0.0 | 1.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.5 | GO:1903426 | regulation of nitric oxide biosynthetic process(GO:0045428) regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 0.7 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.3 | GO:0006490 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 9.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.7 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.2 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.5 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 3.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.4 | 2.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.3 | 0.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 0.7 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.2 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 1.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 0.6 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 1.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.7 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 1.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.7 | GO:0042834 | peptidoglycan binding(GO:0042834) CARD domain binding(GO:0050700) |
| 0.1 | 0.3 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 2.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.1 | GO:0046625 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) sphingolipid binding(GO:0046625) |
| 0.1 | 0.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 1.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 1.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.3 | GO:0008753 | NADPH dehydrogenase (quinone) activity(GO:0008753) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) |
| 0.0 | 0.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.5 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.6 | GO:0001067 | regulatory region DNA binding(GO:0000975) regulatory region nucleic acid binding(GO:0001067) transcription regulatory region DNA binding(GO:0044212) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.2 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | ST_GA12_PATHWAY | G alpha 12 Pathway |