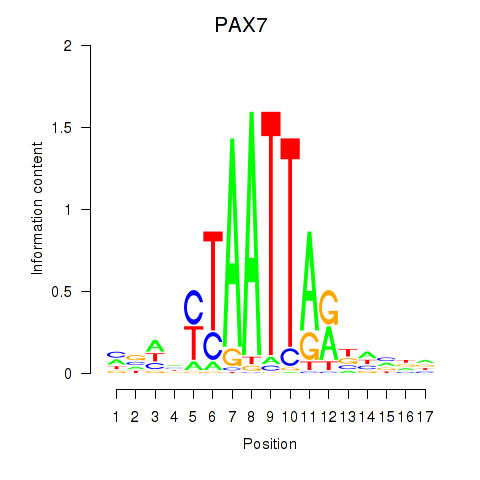
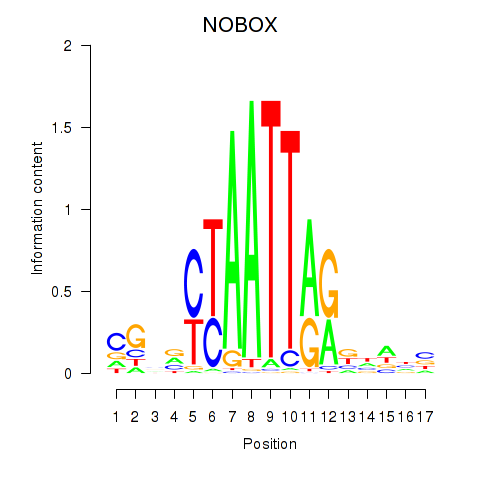
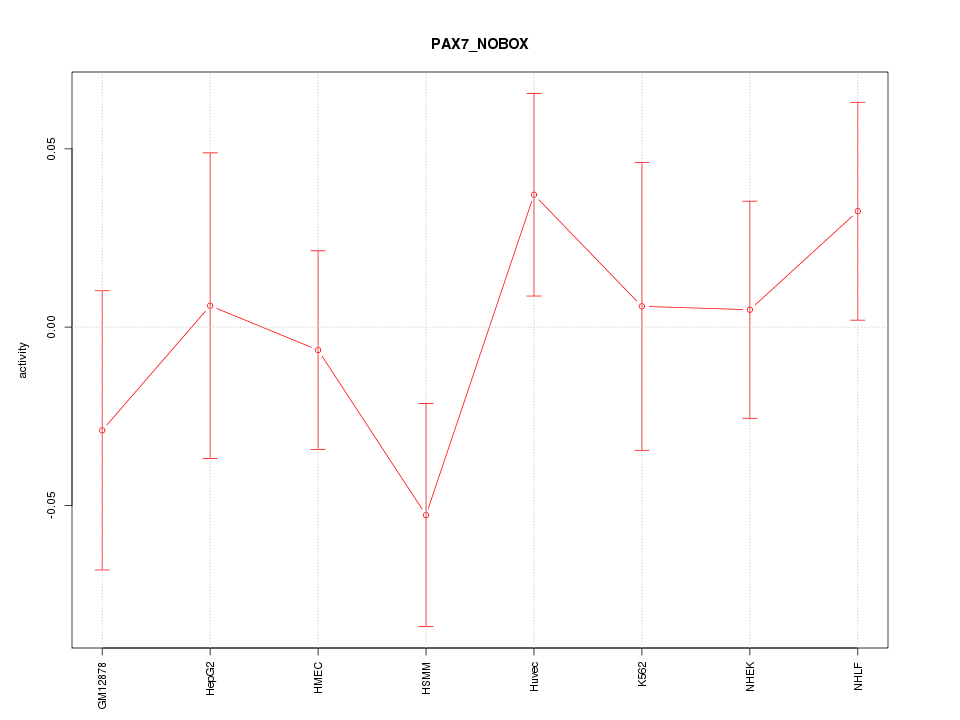
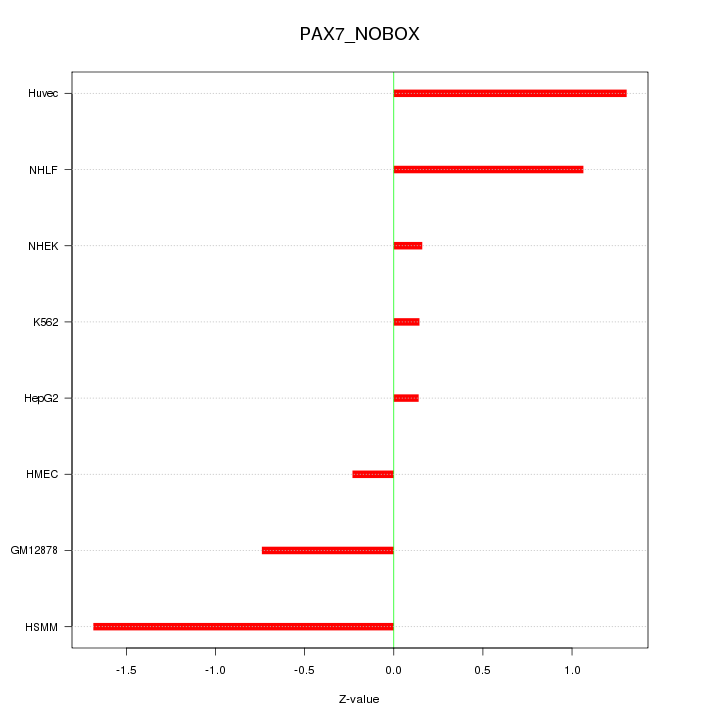
| 0.6 |
1.8 |
GO:0019747 |
regulation of isoprenoid metabolic process(GO:0019747) |
| 0.2 |
0.6 |
GO:0060318 |
definitive erythrocyte differentiation(GO:0060318) |
| 0.2 |
0.7 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 |
1.0 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.1 |
0.4 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.1 |
0.8 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 |
0.6 |
GO:0006264 |
mitochondrial DNA replication(GO:0006264) |
| 0.1 |
0.3 |
GO:0006663 |
platelet activating factor biosynthetic process(GO:0006663) positive regulation of vesicle fusion(GO:0031340) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 |
0.6 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 |
1.3 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
2.4 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
1.9 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.1 |
0.2 |
GO:0060831 |
smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 |
0.3 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.2 |
GO:0001907 |
cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 |
0.2 |
GO:1904358 |
positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 |
0.2 |
GO:0045356 |
positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 |
0.2 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 |
0.3 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 |
0.7 |
GO:0001502 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 |
0.4 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.1 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 |
0.1 |
GO:0042339 |
keratan sulfate metabolic process(GO:0042339) |
| 0.0 |
1.2 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 |
0.2 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 |
0.2 |
GO:0019264 |
L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.0 |
0.1 |
GO:0050955 |
thermoception(GO:0050955) |
| 0.0 |
0.1 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.1 |
GO:0060732 |
positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 |
0.3 |
GO:0048207 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 |
0.2 |
GO:0048488 |
synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.0 |
GO:0051970 |
negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 |
0.1 |
GO:0051919 |
positive regulation of fibrinolysis(GO:0051919) |
| 0.0 |
0.1 |
GO:0072106 |
regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 |
0.1 |
GO:1903332 |
regulation of protein folding(GO:1903332) |
| 0.0 |
0.1 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.0 |
GO:0034126 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) positive regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034126) |
| 0.0 |
0.1 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 |
0.1 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.0 |
0.1 |
GO:0009822 |
alkaloid catabolic process(GO:0009822) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.0 |
GO:0006427 |
histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 |
0.1 |
GO:0032747 |
positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 |
0.0 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
0.4 |
GO:0005980 |
glycogen catabolic process(GO:0005980) |
| 0.0 |
0.0 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 |
0.2 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |