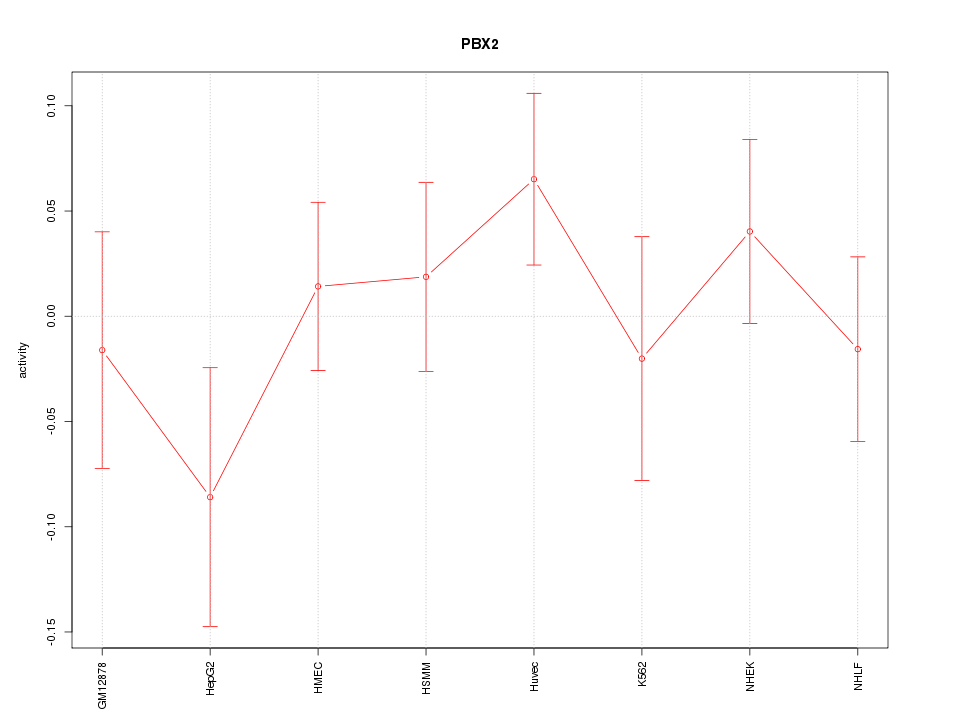
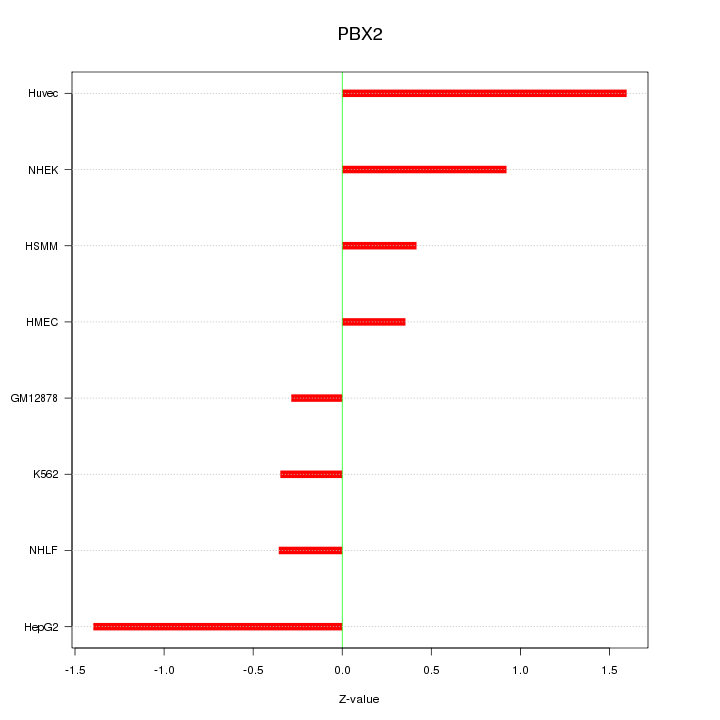
Motif ID: PBX2
Z-value: 0.865
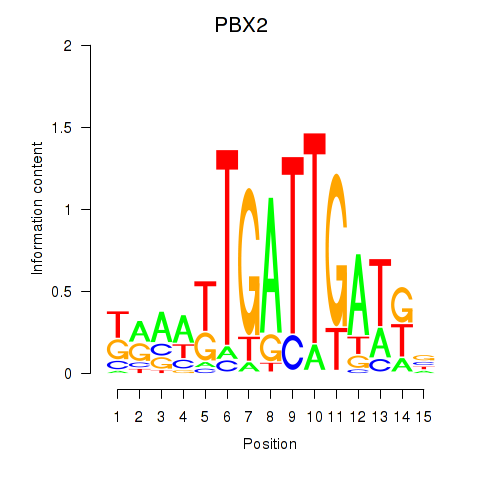
Transcription factors associated with PBX2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PBX2 | ENSG00000204304.7 | PBX2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0060649 | nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.4 | 1.5 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.2 | 0.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 0.5 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.5 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.1 | 0.9 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.4 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 0.2 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.1 | 1.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.4 | GO:0042589 | phagocytic vesicle membrane(GO:0030670) zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.9 | GO:0046625 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) sphingolipid binding(GO:0046625) |
| 0.1 | 1.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.4 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 1.5 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |