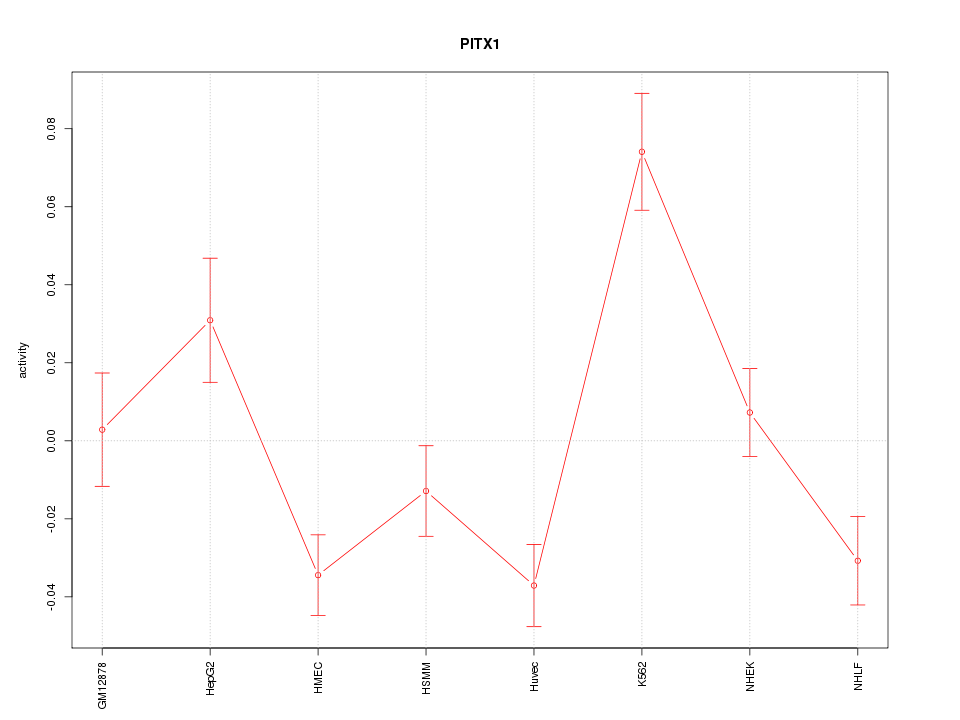
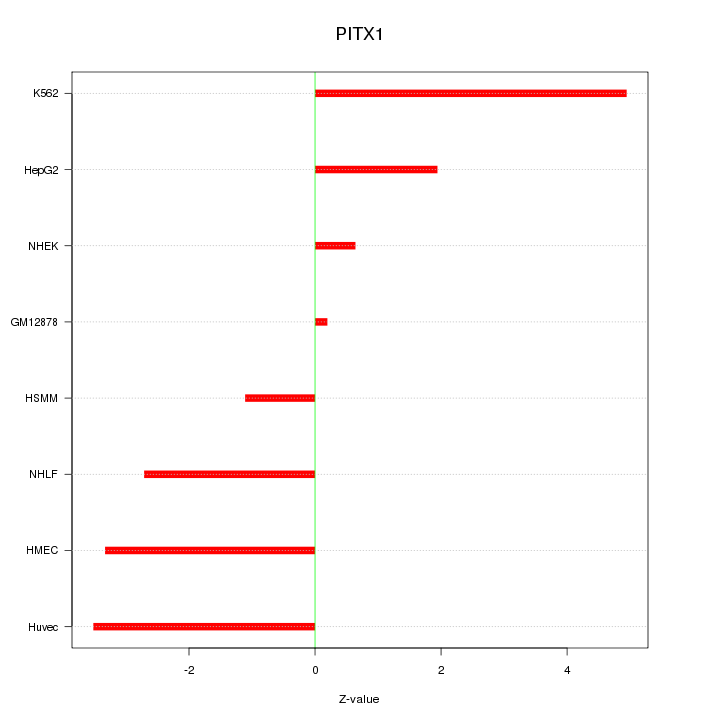
Motif ID: PITX1
Z-value: 2.756
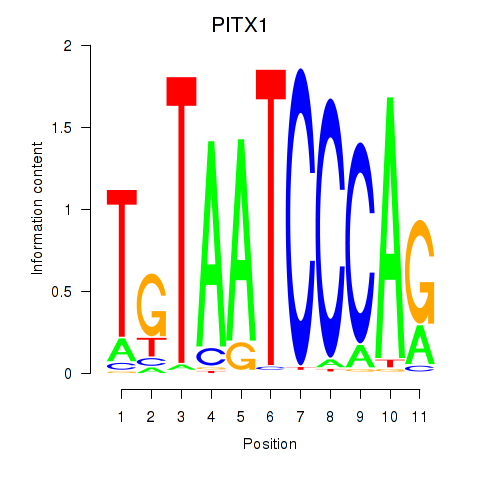
Transcription factors associated with PITX1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PITX1 | ENSG00000069011.11 | PITX1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.9 | 2.6 | GO:2000644 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.8 | 7.6 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.7 | 2.1 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.7 | 2.0 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.7 | 3.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 3.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.6 | 4.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 2.6 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.5 | 1.5 | GO:0097237 | response to cobalt ion(GO:0032025) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.5 | 6.0 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.4 | 0.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.4 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.4 | 3.7 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.4 | 1.2 | GO:0050983 | spermidine catabolic process(GO:0046203) deoxyhypusine biosynthetic process from spermidine(GO:0050983) |
| 0.4 | 8.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.4 | 0.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 | 3.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.4 | 1.1 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.3 | 1.4 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.3 | 1.0 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.3 | 1.0 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) protein hexamerization(GO:0034214) |
| 0.3 | 0.8 | GO:0015854 | guanine transport(GO:0015854) hypoxanthine transport(GO:0035344) thymine transport(GO:0035364) |
| 0.3 | 0.6 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.3 | 1.9 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.3 | 1.3 | GO:0006529 | asparagine metabolic process(GO:0006528) asparagine biosynthetic process(GO:0006529) |
| 0.3 | 1.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 2.0 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.2 | 0.7 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 1.7 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.2 | 1.2 | GO:0008295 | spermidine metabolic process(GO:0008216) spermidine biosynthetic process(GO:0008295) |
| 0.2 | 0.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 1.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.7 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 0.6 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 | 0.6 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 0.6 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.2 | 1.4 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.2 | 2.3 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.2 | 0.6 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 1.5 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.2 | 0.5 | GO:0032242 | regulation of nucleoside transport(GO:0032242) necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.2 | 0.5 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.2 | 0.7 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.2 | 0.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.6 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.2 | 0.5 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 2.0 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.2 | 1.2 | GO:1902624 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.2 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.6 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.1 | 0.4 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.4 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.7 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.1 | 0.5 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.4 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.1 | 1.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.6 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.1 | 0.8 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.6 | GO:0003070 | age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.1 | 2.3 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 1.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 0.9 | GO:0033240 | positive regulation of cellular amine metabolic process(GO:0033240) |
| 0.1 | 0.5 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.1 | 0.5 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.7 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.5 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.1 | 0.8 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.4 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.5 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.6 | GO:0043316 | natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.1 | 1.7 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 1.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 1.5 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.1 | 0.9 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 0.3 | GO:0060744 | thelarche(GO:0042695) development of secondary female sexual characteristics(GO:0046543) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 3.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.2 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.1 | 4.2 | GO:0043038 | tRNA aminoacylation for protein translation(GO:0006418) amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.1 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.5 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.5 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.1 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.4 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.2 | GO:0031077 | post-embryonic camera-type eye development(GO:0031077) post-embryonic organ morphogenesis(GO:0048563) |
| 0.1 | 1.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 2.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.6 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 1.2 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.1 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.4 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.1 | 1.4 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.3 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 1.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.1 | GO:0046668 | retinal cell programmed cell death(GO:0046666) regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 0.6 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 0.4 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 | 0.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.4 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 1.0 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.1 | 0.3 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.9 | GO:0019400 | alditol metabolic process(GO:0019400) |
| 0.1 | 0.5 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 2.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.1 | 0.9 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 4.9 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) ribonucleoprotein complex export from nucleus(GO:0071426) |
| 0.1 | 0.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.7 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.3 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.1 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.2 | GO:0033622 | integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.1 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.5 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 2.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.6 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 1.1 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.1 | 0.3 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.1 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.2 | GO:0030201 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.2 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.4 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.7 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.7 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.0 | 0.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.4 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.5 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 1.2 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:0009128 | AMP catabolic process(GO:0006196) nucleoside monophosphate catabolic process(GO:0009125) purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.4 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 5.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.5 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.6 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.0 | 0.2 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0071474 | cellular water homeostasis(GO:0009992) cellular hyperosmotic response(GO:0071474) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0046368 | GDP-L-fucose biosynthetic process(GO:0042350) 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.1 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.2 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 1.8 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.9 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) nucleoside diphosphate biosynthetic process(GO:0009133) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.6 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.2 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.5 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 3.7 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.3 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.2 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.4 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0030277 | epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 1.1 | GO:0006364 | rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.3 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 1.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.7 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.2 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0060760 | positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.4 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.2 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.9 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) regulation of angiotensin metabolic process(GO:0060177) |
| 0.0 | 0.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.4 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.0 | 0.1 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.2 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 1.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.7 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.3 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.1 | GO:0048703 | embryonic neurocranium morphogenesis(GO:0048702) embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.4 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 2.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.1 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.3 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 1.1 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0032504 | multicellular organism reproduction(GO:0032504) |
| 0.0 | 0.2 | GO:0042462 | photoreceptor cell development(GO:0042461) eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.5 | 3.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 2.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.4 | 2.8 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.3 | 1.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 1.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.3 | 7.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 1.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 3.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 0.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 1.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.9 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.1 | 3.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.9 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.4 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 1.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 4.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.6 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.8 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.9 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 0.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 4.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.5 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 3.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.0 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 5.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.8 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting two-sector ATPase complex(GO:0016469) proton-transporting V-type ATPase complex(GO:0033176) proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 2.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0033177 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 7.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 2.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:1902562 | H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.6 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.4 | GO:0044297 | neuronal cell body(GO:0043025) cell body(GO:0044297) |
| 0.0 | 0.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.2 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 1.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:1990904 | intracellular ribonucleoprotein complex(GO:0030529) ribonucleoprotein complex(GO:1990904) |
| 0.0 | 1.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.0 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.0 | 15.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 0.3 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 1.0 | 5.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.9 | 2.6 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.7 | 2.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.6 | 1.9 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.6 | 8.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.5 | 7.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.5 | 1.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 1.2 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.4 | 2.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.4 | 3.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.4 | 1.4 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.3 | 4.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 1.5 | GO:0080030 | methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.3 | 1.3 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.3 | 0.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.3 | 1.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 0.7 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.2 | 0.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 0.9 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.2 | 1.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 0.8 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 6.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 1.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 0.5 | GO:0031177 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) phosphopantetheine binding(GO:0031177) |
| 0.2 | 0.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.2 | 0.5 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 0.5 | GO:0004917 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) interleukin-2 binding(GO:0019976) |
| 0.1 | 0.6 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.6 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.7 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.4 | GO:0015141 | thiosulfate transmembrane transporter activity(GO:0015117) succinate transmembrane transporter activity(GO:0015141) |
| 0.1 | 0.5 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.1 | 0.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.5 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 3.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.7 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 1.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 1.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 1.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.5 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 5.9 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.3 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.5 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.7 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 0.9 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 1.3 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.1 | 1.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.4 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 0.3 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.6 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 0.3 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 2.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 2.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.5 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.2 | GO:0015145 | monosaccharide transmembrane transporter activity(GO:0015145) |
| 0.1 | 1.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.7 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 0.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.3 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 0.9 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.1 | 1.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.3 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.2 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.1 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.4 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 4.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.8 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.4 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.7 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.1 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 2.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0030611 | arsenate reductase activity(GO:0030611) |
| 0.0 | 1.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.8 | GO:0030528 | obsolete transcription regulator activity(GO:0030528) |
| 0.0 | 0.2 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 1.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.5 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.3 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.4 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.0 | GO:0051213 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) dioxygenase activity(GO:0051213) |
| 0.0 | 0.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 1.0 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 3.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 2.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0016297 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.6 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 5.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 1.9 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.3 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 2.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.5 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 4.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 0.1 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 2.1 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.6 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0060589 | nucleoside-triphosphatase regulator activity(GO:0060589) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.0 | 0.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 5.6 | GO:0003723 | RNA binding(GO:0003723) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.0 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 2.3 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.8 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | ST_GA13_PATHWAY | G alpha 13 Pathway |