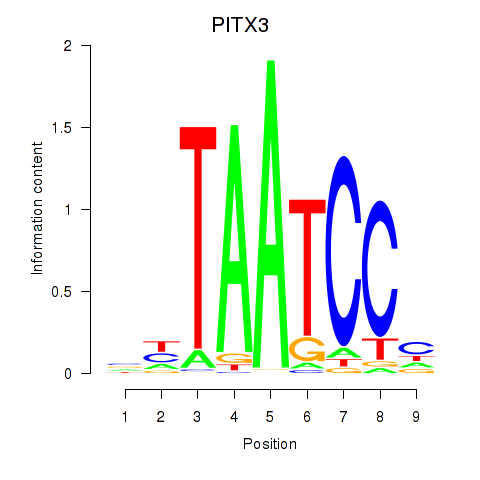
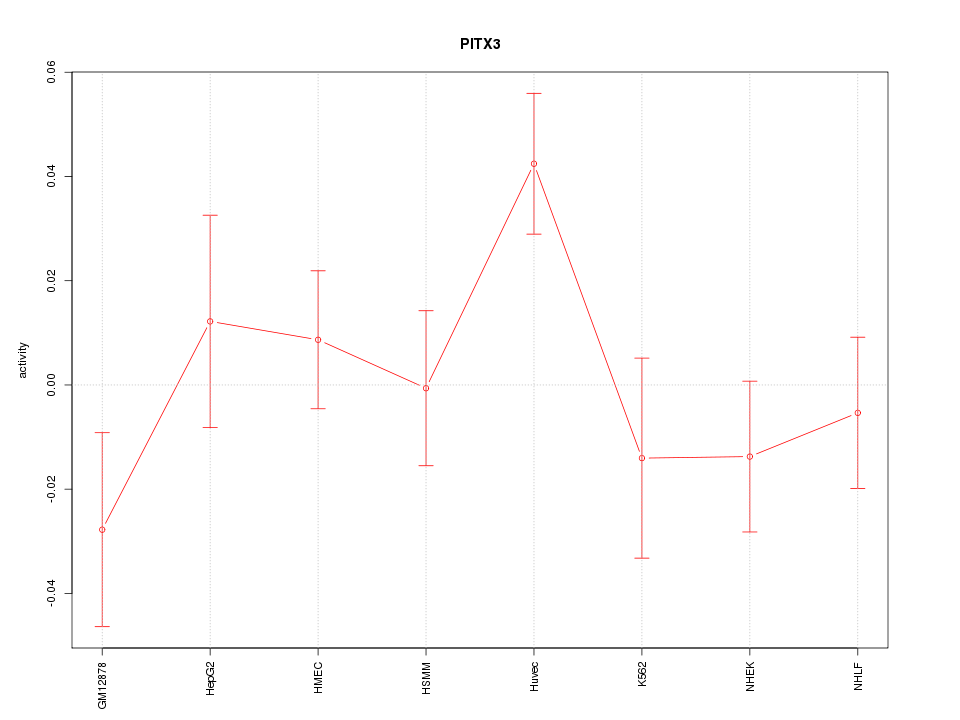
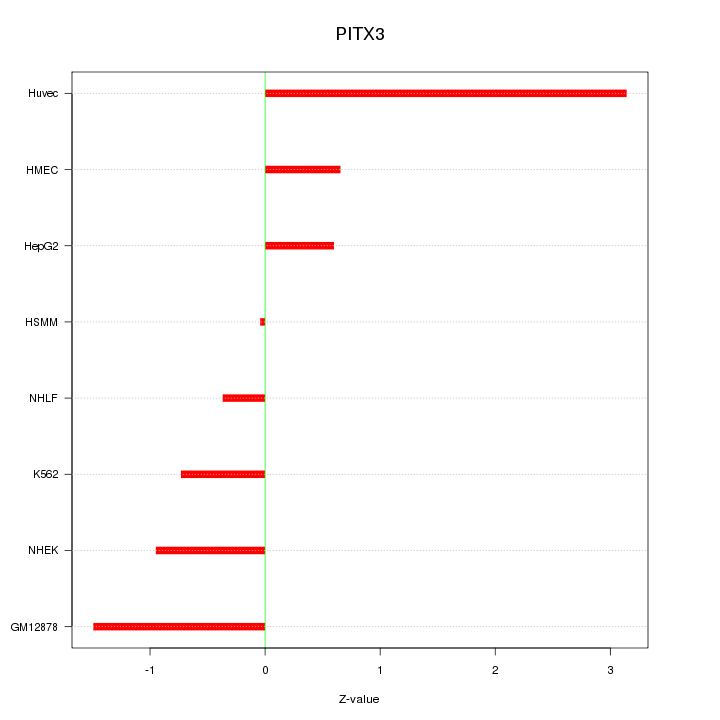
| 0.3 |
1.6 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 |
0.9 |
GO:2000910 |
regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.3 |
1.3 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.3 |
1.2 |
GO:0033986 |
response to methanol(GO:0033986) response to chromate(GO:0046687) |
| 0.3 |
1.8 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 |
1.1 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.3 |
0.8 |
GO:0001757 |
somite specification(GO:0001757) |
| 0.3 |
1.5 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 |
1.0 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.2 |
0.9 |
GO:0001969 |
activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.2 |
1.1 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.2 |
0.6 |
GO:0071380 |
cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 |
0.9 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.2 |
1.1 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.2 |
0.5 |
GO:0006530 |
asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.2 |
1.8 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 |
1.2 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.2 |
0.5 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) |
| 0.2 |
0.7 |
GO:0006154 |
adenosine catabolic process(GO:0006154) |
| 0.2 |
0.5 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.2 |
0.9 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 |
0.6 |
GO:0009956 |
radial pattern formation(GO:0009956) |
| 0.1 |
0.6 |
GO:0034959 |
neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 |
2.3 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 |
2.1 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.1 |
0.4 |
GO:0034036 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 |
0.5 |
GO:0033197 |
response to vitamin E(GO:0033197) |
| 0.1 |
0.3 |
GO:0002124 |
territorial aggressive behavior(GO:0002124) brainstem development(GO:0003360) vocalization behavior(GO:0071625) |
| 0.1 |
0.1 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 |
0.3 |
GO:0046010 |
positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.1 |
0.3 |
GO:0060128 |
corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.1 |
0.6 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 |
0.7 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.1 |
0.3 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 |
0.3 |
GO:0060729 |
negative regulation of interleukin-23 production(GO:0032707) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.6 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.4 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 |
0.3 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.1 |
0.2 |
GO:0071679 |
commissural neuron axon guidance(GO:0071679) |
| 0.1 |
0.2 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
1.7 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.1 |
1.1 |
GO:0007567 |
parturition(GO:0007567) |
| 0.1 |
0.3 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
0.4 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.1 |
0.1 |
GO:0001973 |
adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 |
0.3 |
GO:0015827 |
tryptophan transport(GO:0015827) leucine import(GO:0060356) |
| 0.1 |
0.4 |
GO:0045916 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 |
0.1 |
GO:0032242 |
regulation of nucleoside transport(GO:0032242) |
| 0.1 |
2.7 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
0.1 |
GO:0070244 |
negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.1 |
0.2 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 |
0.2 |
GO:0006726 |
eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 |
0.2 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 |
0.2 |
GO:0022038 |
corpus callosum development(GO:0022038) |
| 0.1 |
0.5 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.1 |
0.2 |
GO:0042339 |
keratan sulfate metabolic process(GO:0042339) |
| 0.1 |
0.4 |
GO:0045780 |
positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 |
0.1 |
GO:0070942 |
neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.0 |
0.1 |
GO:0016554 |
cytidine to uridine editing(GO:0016554) |
| 0.0 |
0.1 |
GO:0010248 |
establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 |
1.2 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
1.1 |
GO:0050927 |
positive regulation of positive chemotaxis(GO:0050927) |
| 0.0 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
1.1 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.0 |
0.1 |
GO:0043254 |
regulation of protein complex assembly(GO:0043254) regulation of cellular component biogenesis(GO:0044087) |
| 0.0 |
0.2 |
GO:0008628 |
hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 |
0.2 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.0 |
1.7 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.5 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 |
0.1 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.1 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 |
0.2 |
GO:0006562 |
proline catabolic process(GO:0006562) |
| 0.0 |
0.1 |
GO:0032621 |
interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 |
0.3 |
GO:0010766 |
negative regulation of sodium ion transport(GO:0010766) |
| 0.0 |
0.3 |
GO:0016199 |
axon midline choice point recognition(GO:0016199) |
| 0.0 |
0.1 |
GO:0008049 |
male courtship behavior(GO:0008049) |
| 0.0 |
0.7 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 |
0.4 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.1 |
GO:1903115 |
regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 |
0.0 |
GO:0030147 |
obsolete natriuresis(GO:0030147) |
| 0.0 |
0.6 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.1 |
GO:0018874 |
benzoate metabolic process(GO:0018874) |
| 0.0 |
0.3 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 |
0.1 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 |
0.2 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.0 |
0.2 |
GO:0010842 |
retina layer formation(GO:0010842) camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 |
0.1 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 |
0.6 |
GO:0006525 |
arginine metabolic process(GO:0006525) |
| 0.0 |
0.5 |
GO:0006693 |
prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 |
0.6 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.2 |
GO:0070243 |
regulation of thymocyte apoptotic process(GO:0070243) |
| 0.0 |
1.2 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.1 |
GO:0008356 |
asymmetric cell division(GO:0008356) |
| 0.0 |
0.3 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.0 |
0.1 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.1 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.0 |
0.1 |
GO:0009296 |
obsolete flagellum assembly(GO:0009296) |
| 0.0 |
0.1 |
GO:0014041 |
regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 |
0.5 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.0 |
0.1 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) embryonic hindgut morphogenesis(GO:0048619) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
0.1 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.2 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.0 |
0.0 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.0 |
0.3 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.1 |
GO:0050955 |
thermoception(GO:0050955) |
| 0.0 |
0.1 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 |
0.1 |
GO:0035269 |
protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 |
0.2 |
GO:0042354 |
fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 |
0.4 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
0.0 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 |
0.1 |
GO:0030029 |
actin filament-based process(GO:0030029) |
| 0.0 |
0.2 |
GO:0010759 |
regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 |
0.1 |
GO:0018027 |
peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 |
0.1 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.0 |
0.2 |
GO:0071391 |
cellular response to estrogen stimulus(GO:0071391) |
| 0.0 |
0.2 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 |
0.2 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
1.0 |
GO:0050728 |
negative regulation of inflammatory response(GO:0050728) |
| 0.0 |
0.1 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.0 |
0.0 |
GO:0071848 |
regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 |
0.0 |
GO:0001999 |
renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) |
| 0.0 |
0.1 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 |
0.1 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.0 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.2 |
GO:0051597 |
response to methylmercury(GO:0051597) |
| 0.0 |
0.4 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.1 |
GO:0034244 |
negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.6 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.1 |
GO:0009136 |
ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 |
0.2 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.0 |
0.1 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 |
0.1 |
GO:0099515 |
nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.0 |
0.4 |
GO:0008211 |
glucocorticoid metabolic process(GO:0008211) |
| 0.0 |
0.0 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.0 |
0.4 |
GO:0048024 |
regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 |
0.1 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) transformation of host cell by virus(GO:0019087) dUMP metabolic process(GO:0046078) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.0 |
0.1 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.0 |
0.2 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.1 |
GO:0030277 |
epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
0.2 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.0 |
0.1 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.7 |
GO:0030837 |
negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 |
0.1 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 |
0.1 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.1 |
GO:0051294 |
establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 |
0.1 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.1 |
GO:0031643 |
regulation of Schwann cell differentiation(GO:0014038) positive regulation of Schwann cell differentiation(GO:0014040) positive regulation of myelination(GO:0031643) |
| 0.0 |
0.2 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.1 |
GO:0042416 |
dopamine biosynthetic process(GO:0042416) |
| 0.0 |
0.1 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.2 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.1 |
GO:0046349 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 |
0.5 |
GO:0044839 |
G2/M transition of mitotic cell cycle(GO:0000086) cell cycle G2/M phase transition(GO:0044839) |
| 0.0 |
0.1 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.4 |
GO:0035338 |
long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 |
0.1 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.1 |
GO:0070669 |
response to interleukin-2(GO:0070669) |
| 0.0 |
0.2 |
GO:0007628 |
adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 |
0.2 |
GO:0000910 |
cytokinesis(GO:0000910) |
| 0.0 |
0.1 |
GO:0006998 |
nuclear envelope organization(GO:0006998) |
| 0.0 |
0.4 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.4 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.3 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 |
0.1 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 |
0.0 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 |
0.5 |
GO:0006023 |
aminoglycan biosynthetic process(GO:0006023) |
| 0.0 |
0.0 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.2 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.0 |
0.1 |
GO:0046668 |
retinal cell programmed cell death(GO:0046666) regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 |
0.2 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.3 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 |
0.1 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) |
| 0.0 |
0.2 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.0 |
0.4 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.0 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.2 |
GO:0048265 |
response to pain(GO:0048265) |
| 0.0 |
0.1 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.2 |
GO:0048207 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 |
0.0 |
GO:0042311 |
vasodilation(GO:0042311) |
| 0.0 |
0.0 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 |
0.1 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 |
0.8 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.0 |
0.0 |
GO:0015864 |
pyrimidine nucleoside transport(GO:0015864) |
| 0.0 |
0.9 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.1 |
GO:0090286 |
nuclear matrix organization(GO:0043578) cytoskeletal anchoring at nuclear membrane(GO:0090286) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.2 |
GO:0009214 |
cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 |
0.0 |
GO:0031282 |
regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 |
0.0 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.0 |
GO:0070265 |
necrotic cell death(GO:0070265) |
| 0.0 |
0.0 |
GO:0009438 |
methylglyoxal metabolic process(GO:0009438) |
| 0.0 |
0.2 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.1 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.0 |
GO:0033089 |
positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 |
0.1 |
GO:0035444 |
nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) ferrous iron transport(GO:0015684) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) ferrous iron import(GO:0070627) iron ion import(GO:0097286) |
| 0.0 |
0.0 |
GO:0043267 |
negative regulation of potassium ion transport(GO:0043267) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.1 |
GO:0051209 |
release of sequestered calcium ion into cytosol(GO:0051209) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) calcium ion transmembrane import into cytosol(GO:0097553) calcium ion import into cytosol(GO:1902656) |
| 0.0 |
0.2 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.1 |
GO:1902591 |
vesicle coating(GO:0006901) COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) single-organism membrane budding(GO:1902591) |
| 0.0 |
0.3 |
GO:0008633 |
obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 |
0.1 |
GO:0021984 |
adenohypophysis development(GO:0021984) |
| 0.0 |
0.0 |
GO:0001692 |
histamine metabolic process(GO:0001692) |
| 0.0 |
0.1 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.0 |
0.0 |
GO:0055075 |
potassium ion homeostasis(GO:0055075) |
| 0.0 |
0.3 |
GO:0019827 |
stem cell population maintenance(GO:0019827) |
| 0.0 |
0.3 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.1 |
GO:0050918 |
positive chemotaxis(GO:0050918) |
| 0.0 |
0.0 |
GO:0070857 |
regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 |
0.9 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.0 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
0.1 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.0 |
GO:0006423 |
cysteinyl-tRNA aminoacylation(GO:0006423) |