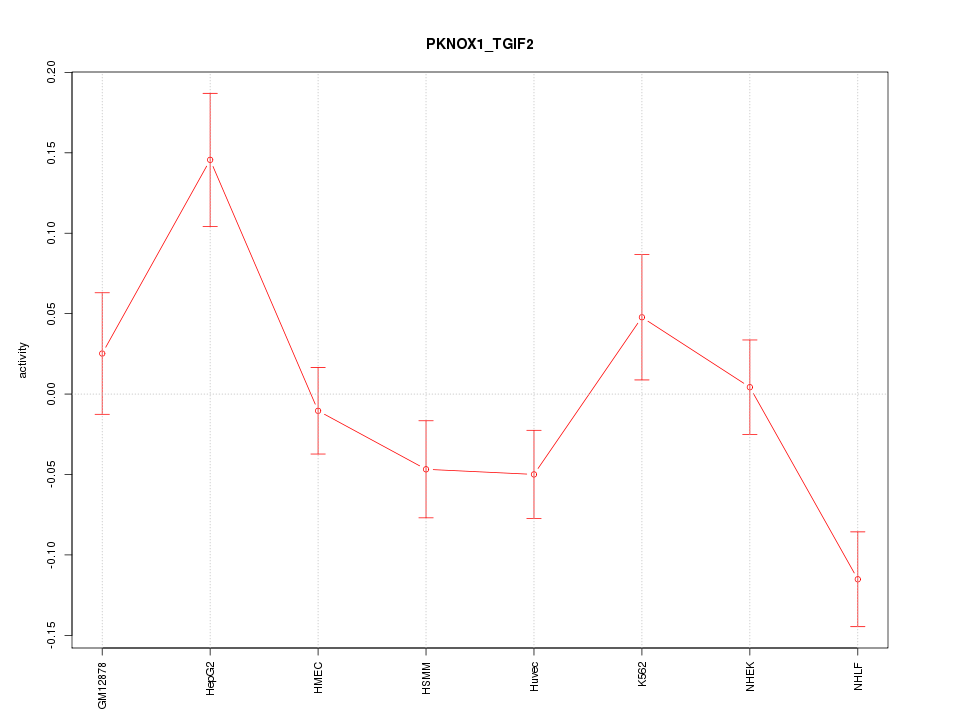
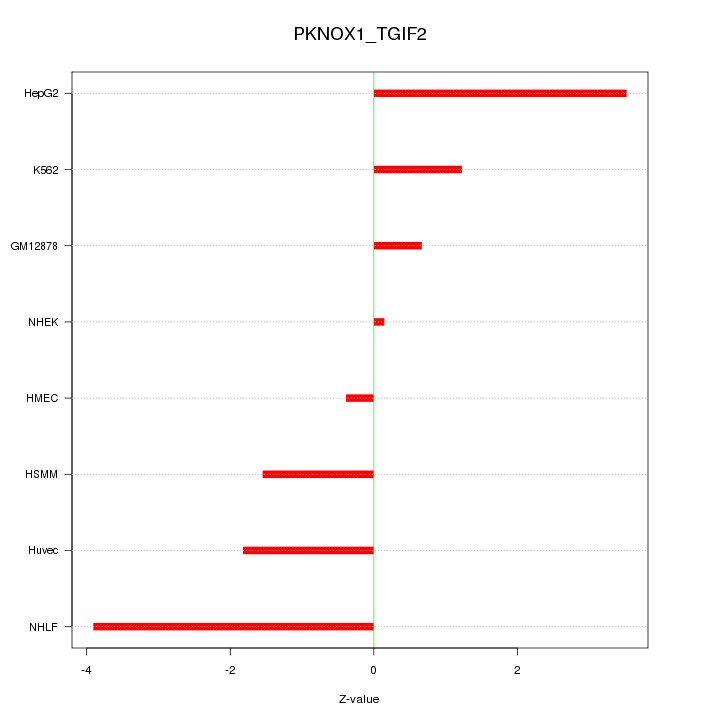
Motif ID: PKNOX1_TGIF2
Z-value: 2.107
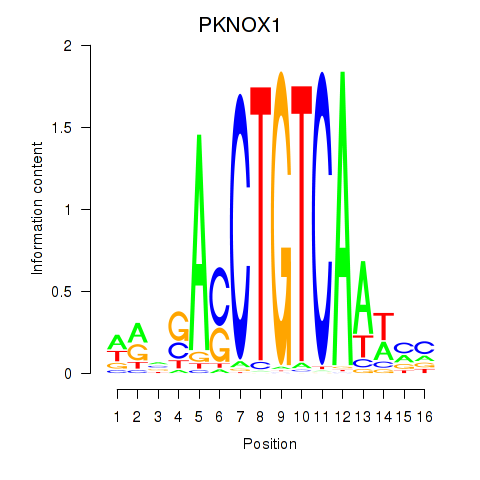
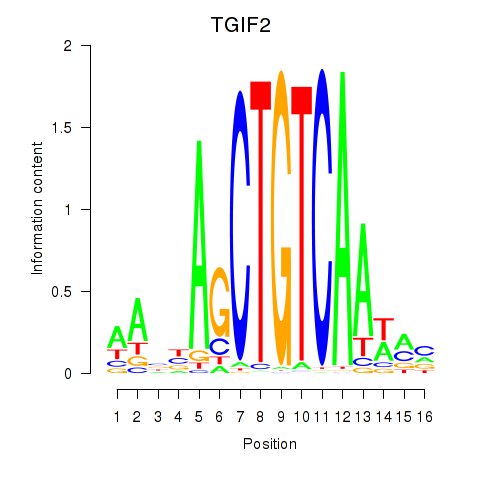
Transcription factors associated with PKNOX1_TGIF2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PKNOX1 | ENSG00000160199.10 | PKNOX1 |
| TGIF2 | ENSG00000118707.5 | TGIF2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.5 | GO:0033986 | response to methanol(GO:0033986) response to chromate(GO:0046687) |
| 1.2 | 3.5 | GO:1901142 | insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 1.0 | 3.1 | GO:0034139 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) regulation of toll-like receptor 3 signaling pathway(GO:0034139) negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.7 | 3.4 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.7 | 2.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.5 | 1.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.4 | 2.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.4 | 5.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.3 | 19.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 1.0 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.3 | 1.3 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) positive regulation of epithelial cell apoptotic process(GO:1904037) regulation of endothelial cell apoptotic process(GO:2000351) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.3 | 1.2 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.3 | 2.4 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.3 | 0.9 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.2 | 0.9 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 1.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 0.8 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 | 1.9 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.2 | 1.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.2 | 2.8 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.2 | 0.7 | GO:0019805 | lactate metabolic process(GO:0006089) quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.7 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 0.5 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.2 | 1.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 1.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.6 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 1.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.9 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.1 | 1.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.6 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 | 1.8 | GO:0009070 | serine family amino acid biosynthetic process(GO:0009070) |
| 0.1 | 0.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.1 | 1.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.9 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.1 | 0.3 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 1.3 | GO:0031055 | DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.1 | GO:0051970 | regulation of transmission of nerve impulse(GO:0051969) negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.9 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.1 | 1.3 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.7 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 2.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.9 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) regulation of cholesterol transporter activity(GO:0060694) |
| 0.0 | 0.3 | GO:1900087 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.8 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.4 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.1 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 0.0 | 1.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0007204 | positive regulation of cytosolic calcium ion concentration(GO:0007204) regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.0 | GO:0050663 | cytokine secretion(GO:0050663) |
| 0.0 | 0.1 | GO:0042754 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian rhythm(GO:0042754) negative regulation of behavior(GO:0048521) |
| 0.0 | 0.8 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 1.1 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 1.6 | GO:0034339 | obsolete regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor(GO:0034339) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.4 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 1.0 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 1.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:1902583 | multi-organism transport(GO:0044766) transport of virus(GO:0046794) intracellular transport of virus(GO:0075733) multi-organism localization(GO:1902579) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.5 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.3 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 8.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.1 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 3.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.9 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 31.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.9 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.8 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 2.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.6 | 1.8 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.5 | 2.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.5 | 2.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 1.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.4 | 1.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 1.3 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.3 | 1.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 0.9 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 11.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.2 | 1.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 0.9 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 1.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 1.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.2 | 0.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 1.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 3.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 0.5 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 1.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.9 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 7.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.3 | GO:0017163 | obsolete basal transcription repressor activity(GO:0017163) |
| 0.1 | 0.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 1.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 2.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 3.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.7 | GO:0030528 | obsolete transcription regulator activity(GO:0030528) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0031177 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) phosphopantetheine binding(GO:0031177) |
| 0.0 | 2.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 0.7 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.6 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.7 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.5 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.0 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.3 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |