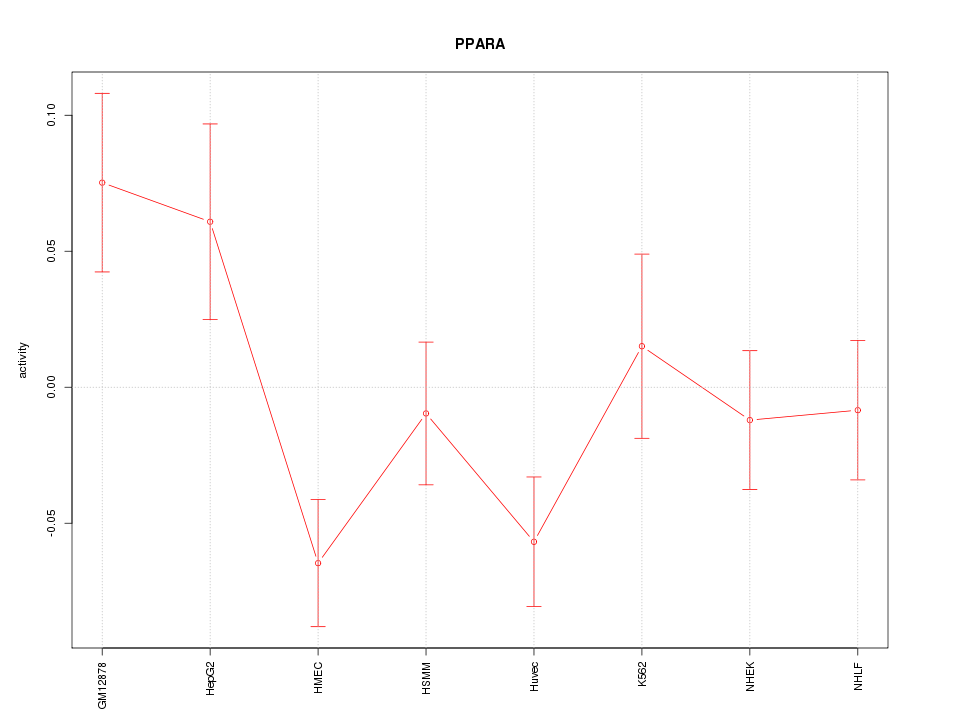
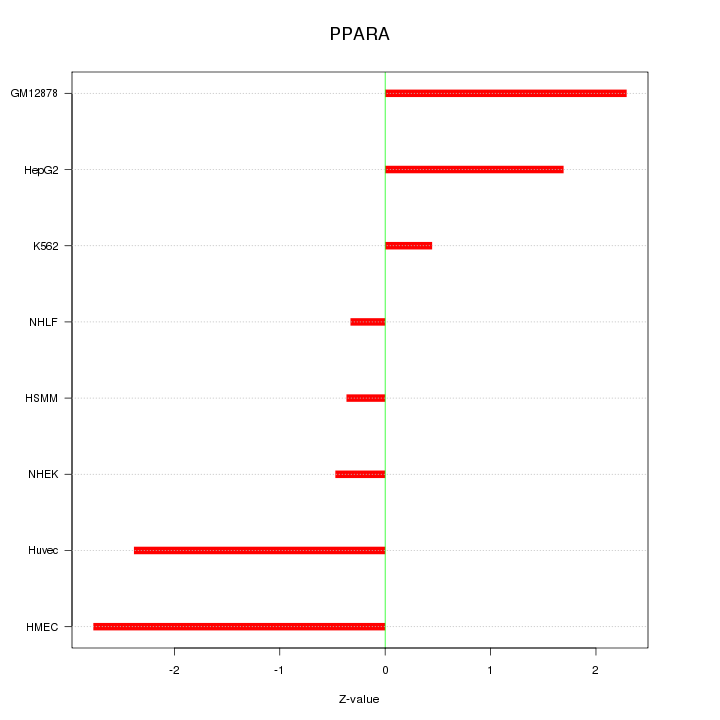
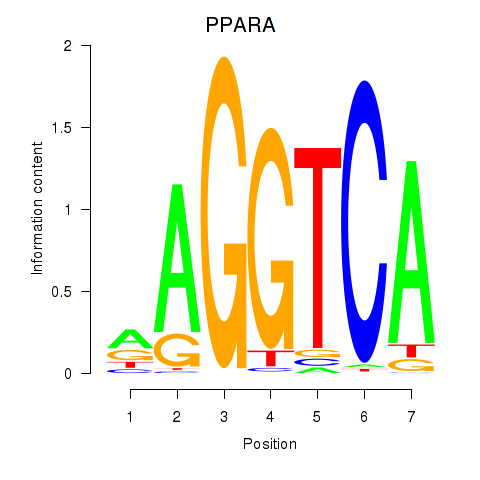
Motif ID: PPARA
Z-value: 1.663
Transcription factors associated with PPARA:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PPARA | ENSG00000186951.12 | PPARA |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0010989 | regulation of high-density lipoprotein particle clearance(GO:0010982) negative regulation of low-density lipoprotein particle clearance(GO:0010989) regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.6 | 2.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.5 | 2.4 | GO:0003093 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.5 | 1.9 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.4 | 2.6 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.4 | 1.6 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.4 | 1.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 0.8 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.3 | 1.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.3 | 0.8 | GO:0060267 | transformation of host cell by virus(GO:0019087) positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 0.8 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.3 | 2.0 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.2 | 0.6 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.2 | 1.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 2.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 0.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 9.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.5 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.2 | 1.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 1.1 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 1.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.4 | GO:0046449 | allantoin metabolic process(GO:0000255) mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.5 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) |
| 0.1 | 0.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.6 | GO:2001259 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.1 | 1.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.1 | 0.4 | GO:2000105 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.1 | 0.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.3 | GO:0007285 | primary spermatocyte growth(GO:0007285) |
| 0.1 | 0.1 | GO:0043270 | positive regulation of ion transport(GO:0043270) |
| 0.1 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 1.0 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 | 0.9 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 1.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.7 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.1 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.4 | GO:0060192 | negative regulation of lipoprotein lipase activity(GO:0051005) negative regulation of lipase activity(GO:0060192) |
| 0.1 | 0.4 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 0.4 | GO:0043501 | skeletal muscle adaptation(GO:0043501) |
| 0.1 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.5 | GO:0015811 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.1 | 0.5 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.8 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.6 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 0.3 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 1.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 0.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) obsolete negative regulation of diuresis(GO:0003077) positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.7 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.3 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.1 | 1.6 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.1 | 0.8 | GO:0002920 | regulation of humoral immune response(GO:0002920) |
| 0.1 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.1 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.2 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.1 | 0.2 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.1 | 0.6 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 | 0.2 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 1.0 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.1 | 0.8 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.1 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.1 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.8 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 1.9 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.2 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.9 | GO:0034764 | positive regulation of transmembrane transport(GO:0034764) positive regulation of ion transmembrane transport(GO:0034767) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0015854 | guanine transport(GO:0015854) hypoxanthine transport(GO:0035344) thymine transport(GO:0035364) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.7 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.5 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 1.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.5 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 1.6 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.5 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.7 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 2.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.2 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 1.0 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.6 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.6 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0043161 | proteasomal protein catabolic process(GO:0010498) proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 2.0 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.2 | GO:0048892 | sensory system development(GO:0048880) lateral line nerve development(GO:0048892) lateral line nerve glial cell differentiation(GO:0048895) lateral line system development(GO:0048925) lateral line nerve glial cell development(GO:0048937) iridophore differentiation(GO:0050935) |
| 0.0 | 0.6 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0034101 | erythrocyte differentiation(GO:0030218) erythrocyte homeostasis(GO:0034101) |
| 0.0 | 0.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.8 | GO:0000080 | mitotic G1 phase(GO:0000080) |
| 0.0 | 0.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.5 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.3 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.5 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 1.3 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 1.2 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0001942 | hair follicle development(GO:0001942) molting cycle process(GO:0022404) hair cycle process(GO:0022405) molting cycle(GO:0042303) hair cycle(GO:0042633) skin epidermis development(GO:0098773) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.6 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.2 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 1.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.5 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.0 | 0.0 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) negative regulation of neuron death(GO:1901215) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.3 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 1.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 2.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 0.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 0.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.5 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.2 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 2.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.8 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 2.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 2.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 1.4 | GO:0035097 | methyltransferase complex(GO:0034708) histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 2.2 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0032421 | stereocilium(GO:0032420) stereocilium bundle(GO:0032421) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 1.7 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.6 | 2.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.5 | 1.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 1.0 | GO:0015141 | thiosulfate transmembrane transporter activity(GO:0015117) succinate transmembrane transporter activity(GO:0015141) |
| 0.3 | 0.9 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.0 | GO:0008907 | integrase activity(GO:0008907) |
| 0.2 | 0.7 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.2 | 2.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 13.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 2.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 0.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 2.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 0.5 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.2 | 0.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 1.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 0.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 1.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 0.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.5 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 2.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.3 | GO:0016531 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.1 | 0.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 1.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.5 | GO:1901567 | icosanoid binding(GO:0050542) icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.1 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.6 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.2 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 2.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.8 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 1.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.3 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 1.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 1.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.0 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.3 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.9 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.5 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.4 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |