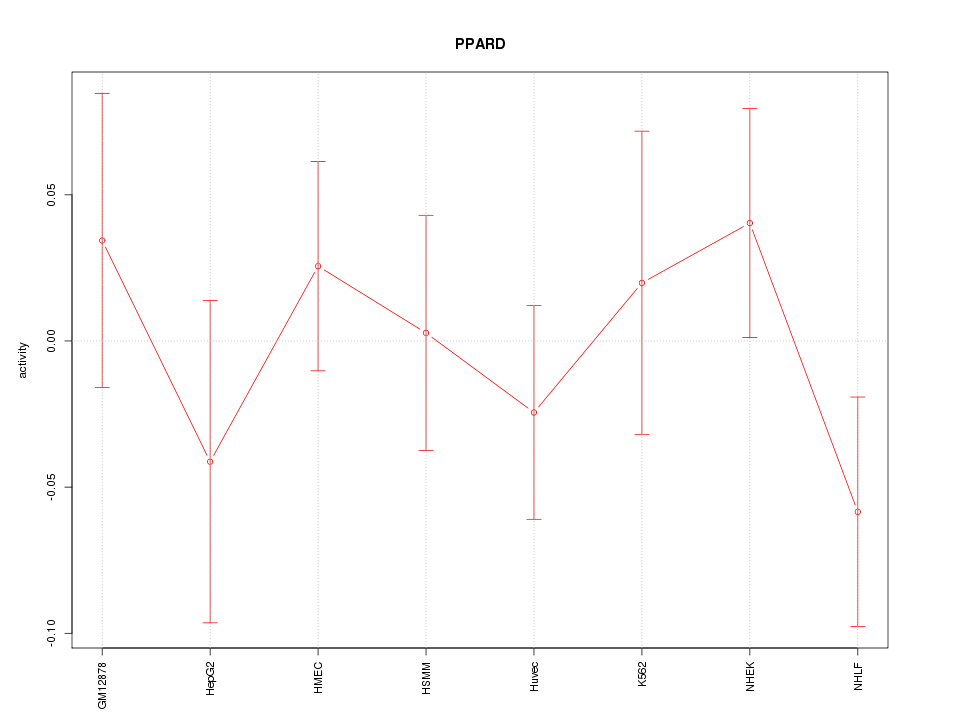
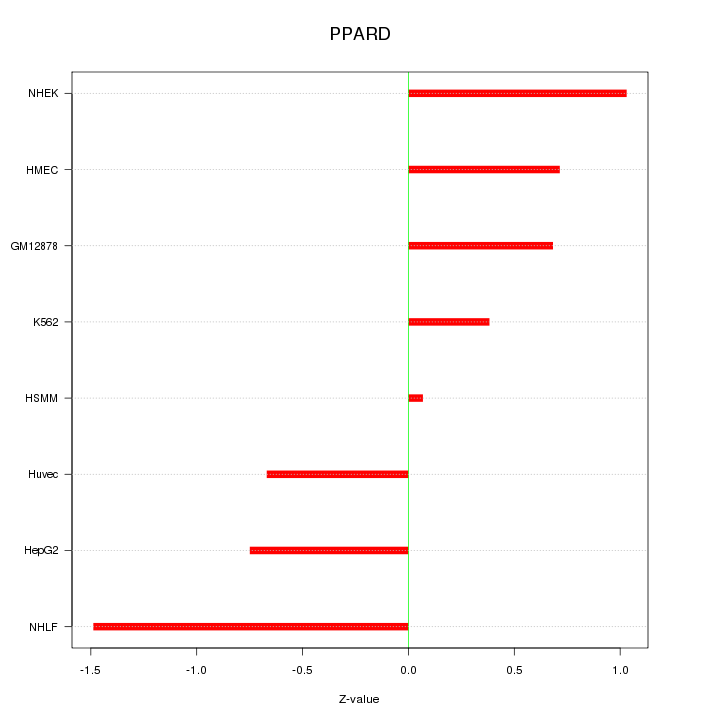
Motif ID: PPARD
Z-value: 0.823
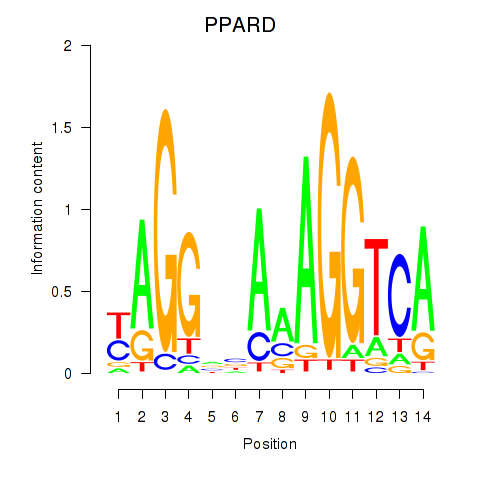
Transcription factors associated with PPARD:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PPARD | ENSG00000112033.9 | PPARD |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) post-embryonic camera-type eye development(GO:0031077) |
| 0.1 | 0.4 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 1.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.9 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.3 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.3 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.2 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.5 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 0.0 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.0 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.4 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0050833 | pyruvate secondary active transmembrane transporter activity(GO:0005477) pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |