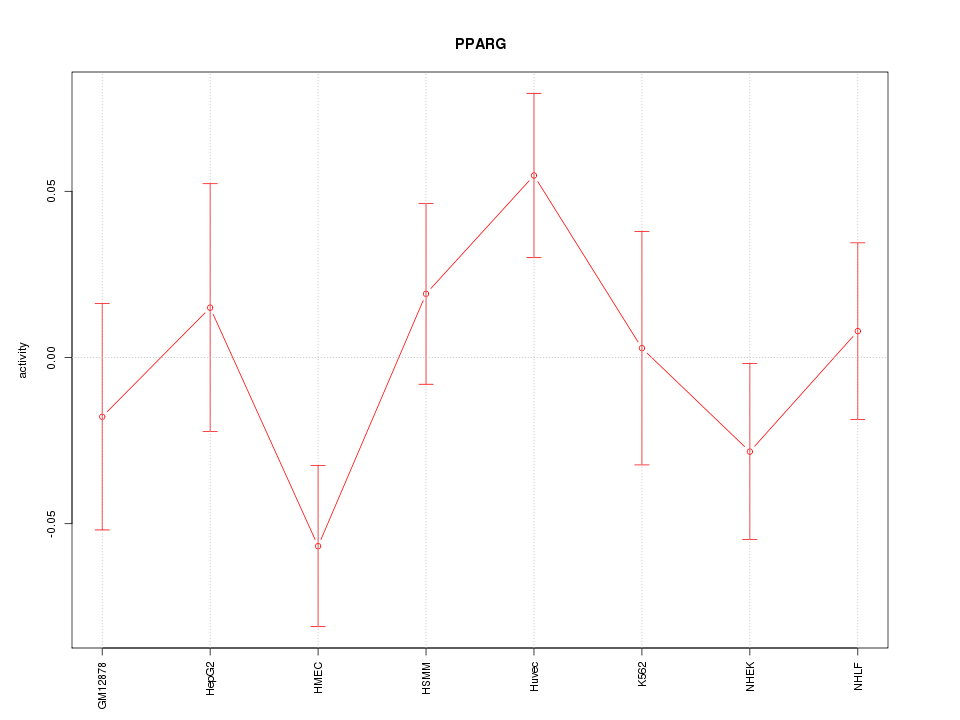
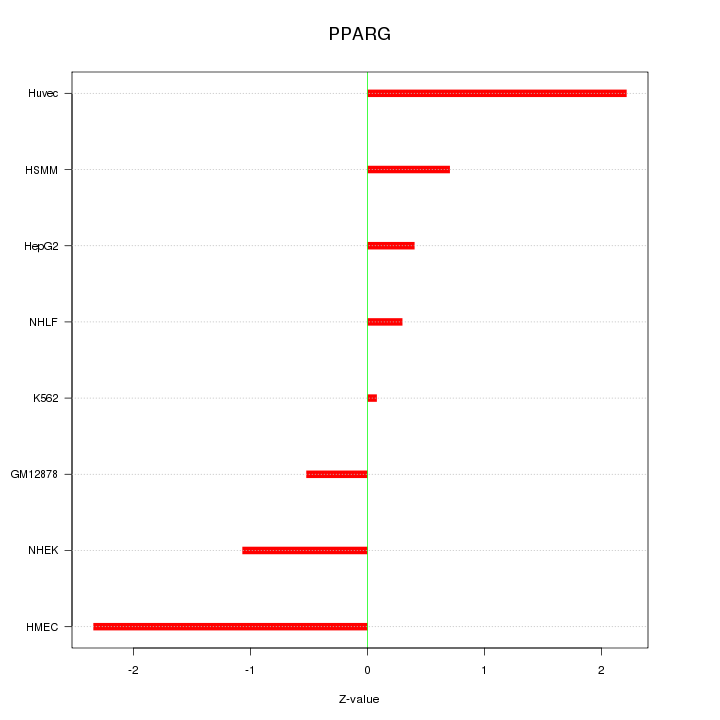
Motif ID: PPARG
Z-value: 1.253
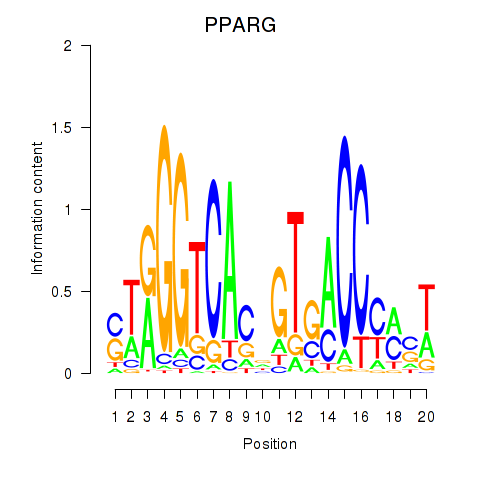
Transcription factors associated with PPARG:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PPARG | ENSG00000132170.15 | PPARG |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0001300 | chronological cell aging(GO:0001300) detection of hypoxia(GO:0070483) |
| 0.2 | 0.8 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.2 | 0.7 | GO:1902116 | negative regulation of organelle assembly(GO:1902116) |
| 0.2 | 1.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 0.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.6 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.4 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.5 | GO:0048170 | optic nerve morphogenesis(GO:0021631) positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 4.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.4 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.1 | 1.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.3 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.1 | 0.3 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.1 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.2 | GO:2000858 | renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) positive regulation of NAD(P)H oxidase activity(GO:0033864) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.0 | 0.1 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 | 0.9 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.1 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.3 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 3.0 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.2 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:2000105 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.0 | 0.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.5 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.4 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.3 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0090103 | outflow tract septum morphogenesis(GO:0003148) cochlea morphogenesis(GO:0090103) |
| 0.0 | 0.1 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.0 | 0.3 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.2 | 2.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.8 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 2.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.7 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.1 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.5 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 1.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.0 | GO:0009279 | cell outer membrane(GO:0009279) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.3 | 0.9 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.2 | 0.6 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 1.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.8 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 4.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.4 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.0 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.2 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |