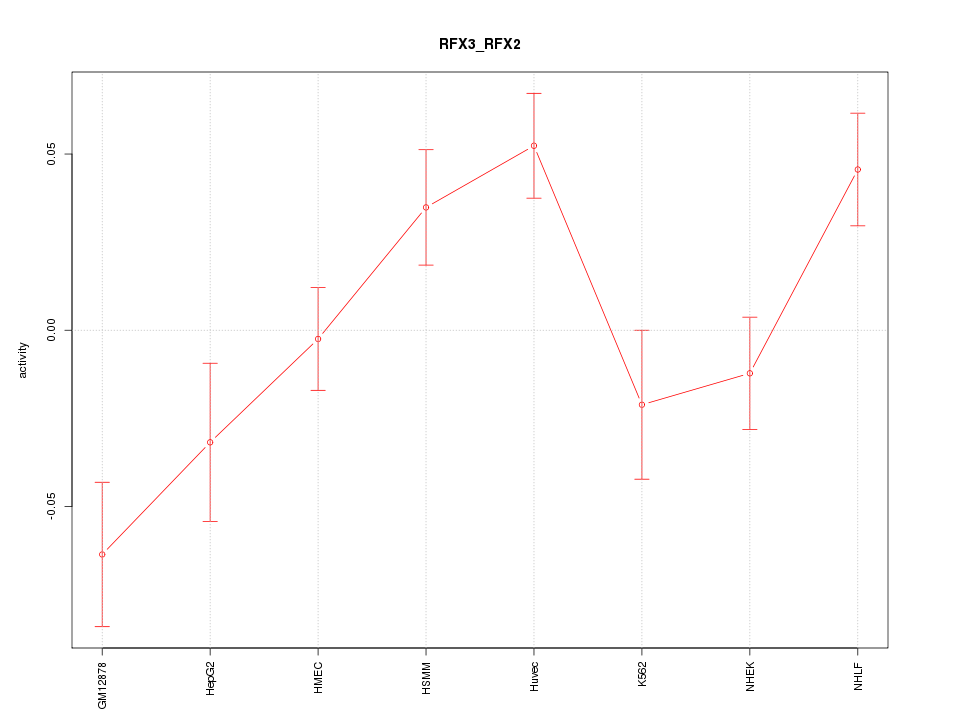
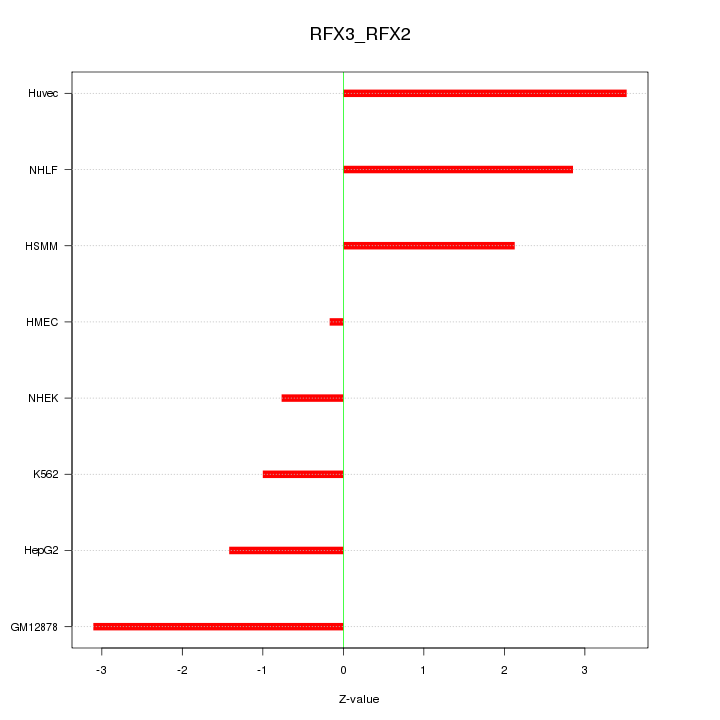
Motif ID: RFX3_RFX2
Z-value: 2.189
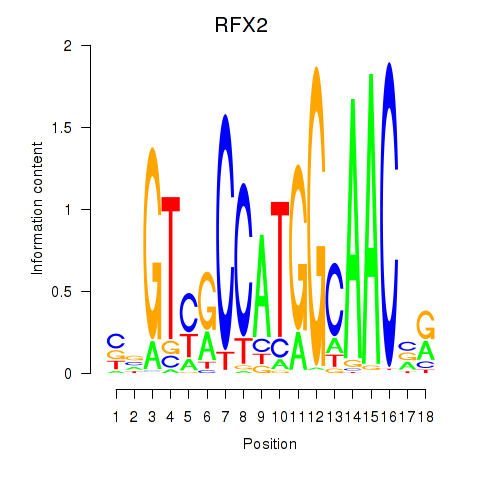
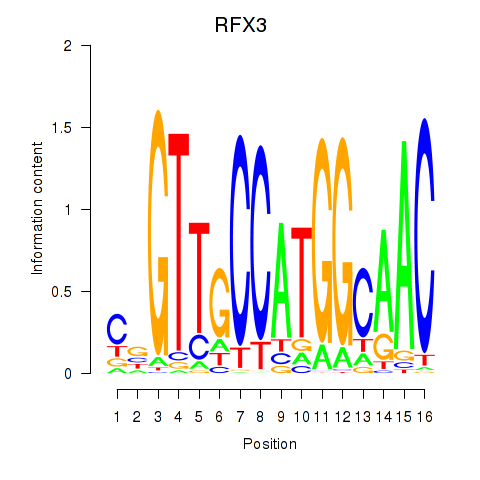
Transcription factors associated with RFX3_RFX2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| RFX2 | ENSG00000087903.8 | RFX2 |
| RFX3 | ENSG00000080298.11 | RFX3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.4 | 1.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.4 | 1.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.4 | 1.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 0.7 | GO:0060295 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.3 | 1.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.3 | 0.9 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 | 0.3 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.3 | 0.8 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.3 | 1.3 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.2 | 3.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.2 | 2.5 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.2 | 0.9 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 1.5 | GO:0046349 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 1.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 0.8 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.2 | 0.2 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.2 | 1.5 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 | 1.6 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.2 | 2.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 4.1 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 1.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 2.5 | GO:0042517 | positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.1 | 0.5 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 1.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 1.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.4 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.1 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.5 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.2 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.5 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 2.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 0.7 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 2.3 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.1 | 0.4 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 3.4 | GO:1900181 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein localization to nucleus(GO:1900181) negative regulation of protein import(GO:1904590) |
| 0.1 | 0.2 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 0.2 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 0.4 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.1 | 0.2 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.1 | 1.0 | GO:0003407 | neural retina development(GO:0003407) |
| 0.1 | 1.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.8 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 11.7 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 1.1 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 3.0 | GO:0008064 | regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.0 | 0.6 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 1.2 | GO:0055007 | cardiac muscle cell differentiation(GO:0055007) |
| 0.0 | 2.3 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.6 | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
| 0.0 | 0.7 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 8.1 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.2 | GO:0016559 | peroxisome membrane biogenesis(GO:0016557) peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.9 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.5 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.8 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.9 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 1.3 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.7 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.3 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 | 1.1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.0 | 5.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) actin-mediated cell contraction(GO:0070252) |
| 0.0 | 0.1 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.8 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.1 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.3 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.3 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.5 | GO:0007569 | cell aging(GO:0007569) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 1.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 2.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.2 | 3.2 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.1 | 3.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 4.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 4.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 4.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 2.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 2.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 3.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 4.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.4 | 1.5 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.3 | 4.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 0.9 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 2.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.6 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.2 | 1.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 1.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 2.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 2.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 1.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.8 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.1 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.6 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 8.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.7 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 1.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 1.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 3.3 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 2.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 2.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.1 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 2.9 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.0 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |