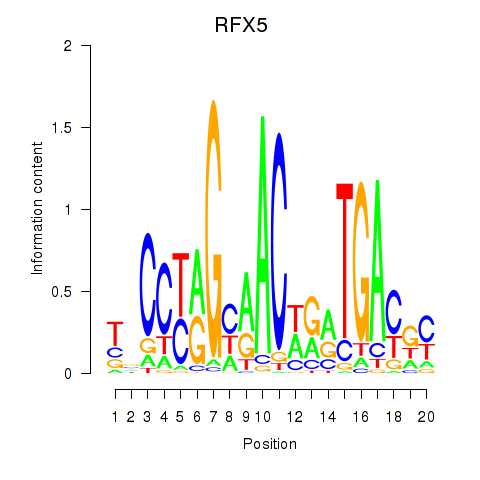
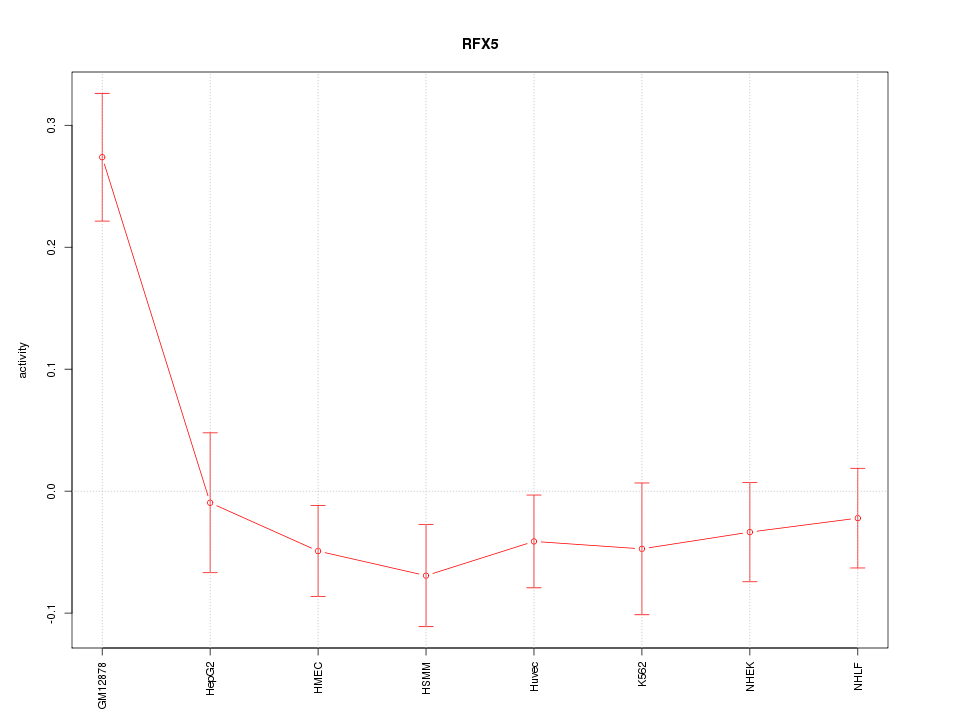
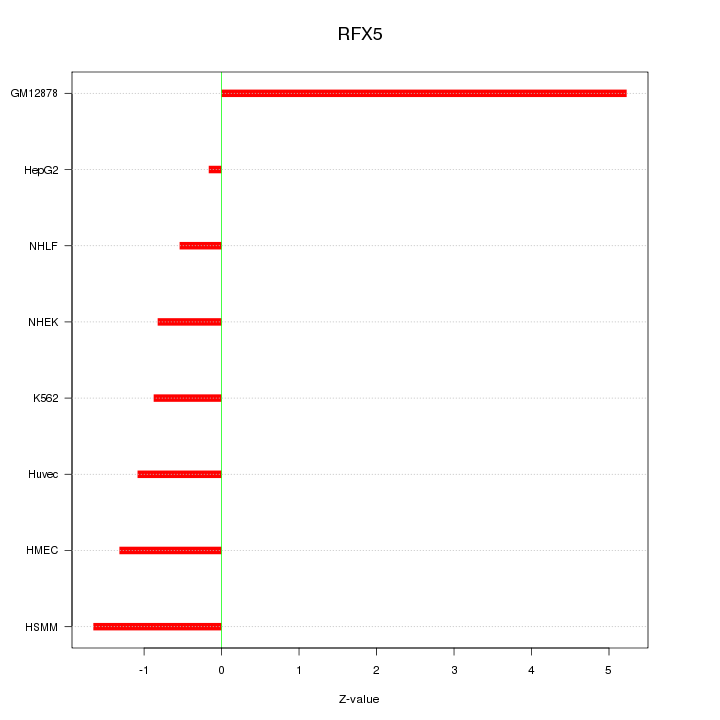
|
chr6_-_32557610
|
9.727
|
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1
|
|
chr6_-_32498046
|
8.990
|
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr5_-_149792295
|
8.279
|
ENST00000518797.1
ENST00000524315.1
ENST00000009530.7
ENST00000377795.3
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr6_+_33043703
|
8.207
|
ENST00000418931.2
ENST00000535465.1
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr6_-_32920794
|
7.437
|
ENST00000395305.3
ENST00000395303.3
ENST00000374843.4
ENST00000429234.1
|
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha
Uncharacterized protein
|
|
chr6_-_32908792
|
6.435
|
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr6_-_32634425
|
5.747
|
ENST00000399082.3
ENST00000399079.3
ENST00000374943.4
ENST00000434651.2
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr6_+_32821924
|
4.770
|
ENST00000374859.2
ENST00000453265.2
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9
|
|
chr6_-_32784687
|
4.502
|
ENST00000447394.1
ENST00000438763.2
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr6_+_32812568
|
4.063
|
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9
|
|
chr6_-_32812420
|
3.793
|
ENST00000374881.2
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8
|
|
chr6_-_32731299
|
3.212
|
ENST00000435145.2
ENST00000437316.2
|
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2
|
|
chr13_-_47012325
|
3.074
|
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like
|
|
chr6_-_32821599
|
2.880
|
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr2_-_242088850
|
2.683
|
ENST00000358649.4
ENST00000452907.1
ENST00000403638.3
ENST00000539818.1
ENST00000415234.1
ENST00000234040.4
|
PASK
|
PAS domain containing serine/threonine kinase
|
|
chr6_-_32908765
|
2.598
|
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr1_+_181003067
|
1.608
|
ENST00000434571.2
ENST00000367579.3
ENST00000282990.6
ENST00000367580.5
|
MR1
|
major histocompatibility complex, class I-related
|
|
chr6_-_32731243
|
1.505
|
ENST00000427449.1
ENST00000411527.1
|
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2
|
|
chr6_+_26383404
|
1.366
|
ENST00000416795.2
ENST00000494184.1
|
BTN2A2
|
butyrophilin, subfamily 2, member A2
|
|
chr6_+_26383318
|
1.295
|
ENST00000469230.1
ENST00000490025.1
ENST00000356709.4
ENST00000352867.2
ENST00000493275.1
ENST00000472507.1
ENST00000482536.1
ENST00000432533.2
ENST00000482842.1
|
BTN2A2
|
butyrophilin, subfamily 2, member A2
|
|
chr1_+_249132462
|
1.271
|
ENST00000306562.3
|
ZNF672
|
zinc finger protein 672
|
|
chr6_-_133079022
|
0.960
|
ENST00000525289.1
ENST00000326499.6
|
VNN2
|
vanin 2
|
|
chr19_+_36103631
|
0.905
|
ENST00000203166.5
ENST00000379045.2
|
HAUS5
|
HAUS augmin-like complex, subunit 5
|
|
chr16_+_67381263
|
0.843
|
ENST00000541146.1
ENST00000563189.1
ENST00000290940.7
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr6_+_26440700
|
0.834
|
ENST00000494393.1
ENST00000482451.1
ENST00000244519.2
ENST00000339789.4
ENST00000471353.1
ENST00000361232.3
ENST00000487627.1
ENST00000496719.1
ENST00000490254.1
ENST00000487272.1
|
BTN3A3
|
butyrophilin, subfamily 3, member A3
|
|
chr8_-_98290087
|
0.829
|
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5
|
|
chr16_+_67381289
|
0.790
|
ENST00000435835.3
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr16_-_74808710
|
0.782
|
ENST00000219368.3
ENST00000544337.1
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr2_+_242089046
|
0.651
|
ENST00000407025.1
ENST00000272983.8
|
PPP1R7
|
protein phosphatase 1, regulatory subunit 7
|
|
chr2_+_242088963
|
0.561
|
ENST00000438799.1
ENST00000402734.1
ENST00000423280.1
|
PPP1R7
|
protein phosphatase 1, regulatory subunit 7
|
|
chr8_+_101170257
|
0.444
|
ENST00000251809.3
|
SPAG1
|
sperm associated antigen 1
|
|
chr6_+_26365443
|
0.398
|
ENST00000527422.1
ENST00000356386.2
ENST00000396934.3
ENST00000377708.2
ENST00000396948.1
ENST00000508906.2
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr4_-_83719884
|
0.365
|
ENST00000282709.4
ENST00000273908.4
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr20_+_42875935
|
0.338
|
ENST00000438466.1
ENST00000372952.3
ENST00000537864.1
ENST00000445952.1
|
GDAP1L1
|
ganglioside induced differentiation associated protein 1-like 1
|
|
chr20_+_42875887
|
0.301
|
ENST00000342560.5
|
GDAP1L1
|
ganglioside induced differentiation associated protein 1-like 1
|
|
chr6_-_24666819
|
0.264
|
ENST00000341060.3
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2
|
|
chr2_+_173600565
|
0.261
|
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr2_+_173600514
|
0.258
|
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr6_-_24667180
|
0.104
|
ENST00000545995.1
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2
|
|
chr8_-_99306611
|
0.092
|
ENST00000341166.3
|
NIPAL2
|
NIPA-like domain containing 2
|
|
chr3_+_196466710
|
0.085
|
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr8_-_99306564
|
0.053
|
ENST00000430223.2
|
NIPAL2
|
NIPA-like domain containing 2
|
|
chr6_-_30181133
|
0.046
|
ENST00000454678.2
ENST00000434785.1
|
TRIM26
|
tripartite motif containing 26
|
|
chr1_+_36549676
|
0.045
|
ENST00000207457.3
|
TEKT2
|
tektin 2 (testicular)
|
|
chr6_-_30181156
|
0.010
|
ENST00000418026.1
ENST00000416596.1
ENST00000453195.1
|
TRIM26
|
tripartite motif containing 26
|