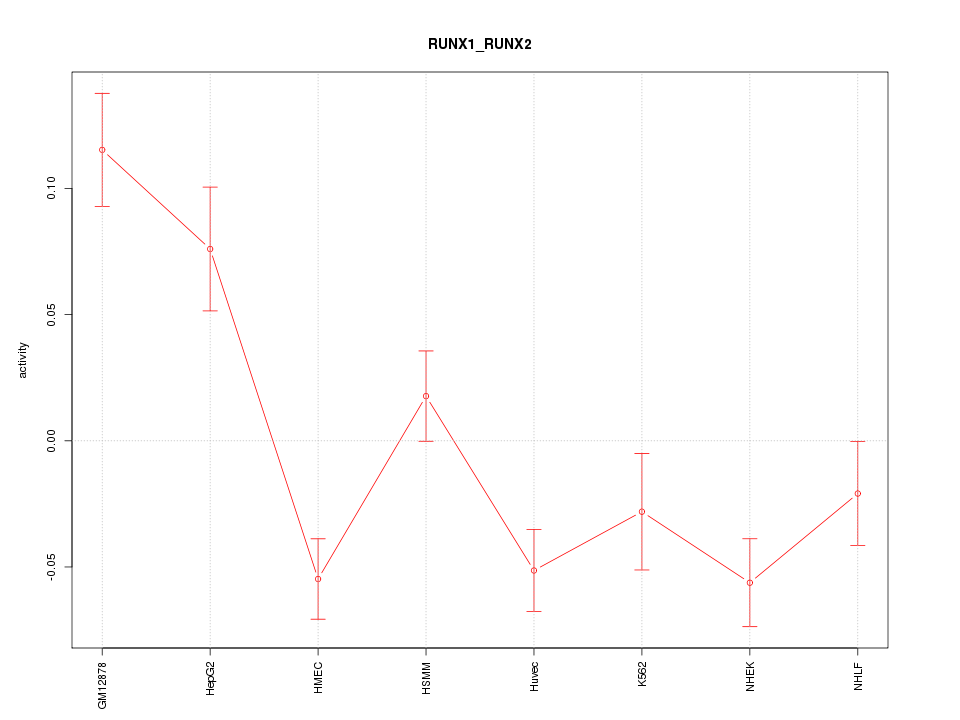
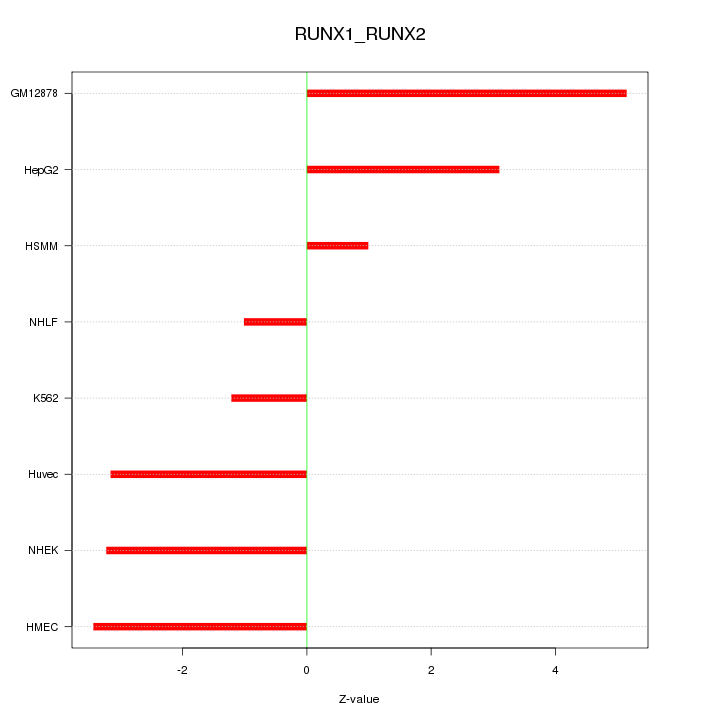
Motif ID: RUNX1_RUNX2
Z-value: 2.996
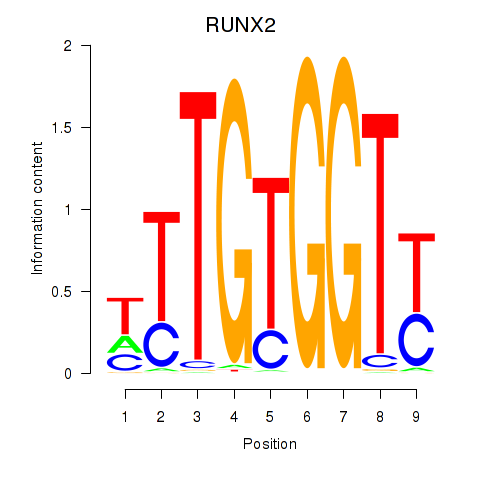
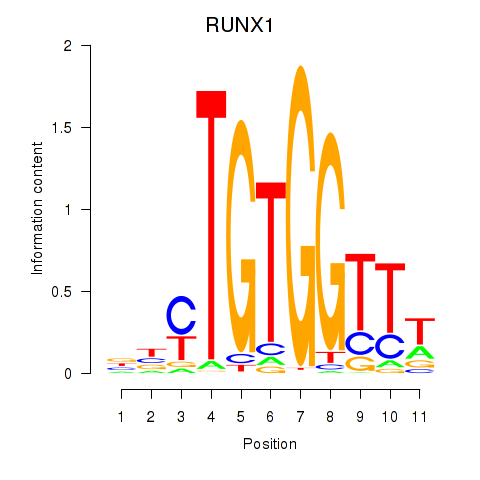
Transcription factors associated with RUNX1_RUNX2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| RUNX1 | ENSG00000159216.14 | RUNX1 |
| RUNX2 | ENSG00000124813.16 | RUNX2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 1.3 | 5.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 1.2 | 3.5 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.7 | 2.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.7 | 2.1 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.6 | 1.9 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.6 | 1.9 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.6 | 2.5 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.6 | 1.8 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.6 | 4.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.6 | 2.3 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.5 | 5.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.5 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.5 | 2.0 | GO:0033622 | integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.5 | 1.4 | GO:0001907 | cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.5 | 2.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.4 | 4.9 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.4 | 1.6 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.4 | 1.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.4 | 0.8 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.4 | 1.4 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.3 | 0.7 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 1.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.3 | 1.7 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.3 | 1.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 1.0 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.3 | 0.9 | GO:0048143 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) astrocyte activation(GO:0048143) |
| 0.3 | 3.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 7.6 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.3 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 0.6 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.3 | 4.8 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.3 | 2.7 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.3 | 4.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 1.6 | GO:0070168 | negative regulation of bone mineralization(GO:0030502) negative regulation of biomineral tissue development(GO:0070168) |
| 0.3 | 4.7 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.3 | 0.8 | GO:0046210 | nitric oxide catabolic process(GO:0046210) |
| 0.3 | 1.0 | GO:0033986 | response to methanol(GO:0033986) response to chromate(GO:0046687) |
| 0.2 | 0.2 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 0.2 | 1.7 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.2 | 0.9 | GO:0002682 | regulation of immune system process(GO:0002682) |
| 0.2 | 0.6 | GO:0003289 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.2 | 0.8 | GO:0050653 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 1.2 | GO:0099515 | nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.2 | 0.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.6 | GO:0010430 | ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 0.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 1.5 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.2 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 0.9 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.2 | 1.4 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.2 | 0.2 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 8.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 2.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 3.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 3.5 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.1 | 0.4 | GO:1990170 | detoxification of cadmium ion(GO:0071585) stress response to cadmium ion(GO:1990170) |
| 0.1 | 0.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 4.1 | GO:0045069 | regulation of viral genome replication(GO:0045069) |
| 0.1 | 0.6 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 1.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.1 | 0.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 1.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.5 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 1.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.3 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.5 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.1 | 1.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.6 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 1.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.4 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.8 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 1.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.7 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 3.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.3 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 1.8 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 0.4 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.1 | 0.3 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.1 | 1.9 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 1.4 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.6 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.3 | GO:2000858 | renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) positive regulation of NAD(P)H oxidase activity(GO:0033864) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.1 | 0.5 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.7 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.4 | GO:0001941 | postsynaptic membrane organization(GO:0001941) |
| 0.1 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.1 | 0.3 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.6 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 1.5 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.1 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.7 | GO:0060323 | head morphogenesis(GO:0060323) |
| 0.1 | 0.5 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.1 | 0.4 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.6 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.5 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 1.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 1.9 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.1 | 0.5 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.1 | 3.3 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.1 | 0.5 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.1 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.5 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.1 | 1.7 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.1 | 1.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.3 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 5.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.5 | GO:0060042 | retina morphogenesis in camera-type eye(GO:0060042) |
| 0.1 | 0.2 | GO:0007285 | primary spermatocyte growth(GO:0007285) |
| 0.1 | 0.2 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) positive regulation of ossification(GO:0045778) |
| 0.1 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.5 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.3 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 0.3 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 1.7 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.1 | 0.1 | GO:0070849 | response to epidermal growth factor(GO:0070849) |
| 0.1 | 0.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.2 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 0.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 1.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.4 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.1 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.4 | GO:0043954 | cell junction maintenance(GO:0034331) cellular component maintenance(GO:0043954) cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.2 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 0.8 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.2 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.1 | 3.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.9 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.4 | GO:0006984 | ER-nucleus signaling pathway(GO:0006984) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.7 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.6 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 2.2 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:0014831 | intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 3.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 3.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.8 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 2.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.7 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.1 | GO:0016071 | mRNA metabolic process(GO:0016071) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.7 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 2.2 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.2 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 2.9 | GO:0007204 | positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 | 3.9 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.5 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 2.1 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 4.9 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.3 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 1.9 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.6 | GO:0071593 | T cell activation(GO:0042110) leukocyte aggregation(GO:0070486) T cell aggregation(GO:0070489) lymphocyte aggregation(GO:0071593) |
| 0.0 | 0.1 | GO:0045953 | negative regulation of natural killer cell mediated immunity(GO:0002716) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.0 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleotide catabolic process(GO:0009155) deoxyribonucleoside triphosphate catabolic process(GO:0009204) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.5 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0030578 | nuclear body organization(GO:0030575) PML body organization(GO:0030578) |
| 0.0 | 0.0 | GO:0070474 | uterine smooth muscle contraction(GO:0070471) regulation of uterine smooth muscle contraction(GO:0070472) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 0.1 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.3 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.1 | GO:0032414 | positive regulation of ion transmembrane transporter activity(GO:0032414) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.9 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.2 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.2 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0003279 | cardiac septum development(GO:0003279) |
| 0.0 | 0.1 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.0 | 0.2 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.0 | GO:0002252 | immune effector process(GO:0002252) |
| 0.0 | 0.8 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.1 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.0 | 0.1 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) |
| 0.0 | 0.1 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.1 | GO:0046718 | entry into host cell(GO:0030260) entry into host(GO:0044409) viral entry into host cell(GO:0046718) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.0 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) chondrocyte development(GO:0002063) |
| 0.0 | 0.7 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.0 | GO:0002124 | aggressive behavior(GO:0002118) territorial aggressive behavior(GO:0002124) brainstem development(GO:0003360) vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0042168 | heme metabolic process(GO:0042168) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.6 | 1.8 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.5 | 1.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.5 | 2.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.5 | 3.3 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.4 | 7.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 1.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 1.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 4.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 2.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 0.9 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 1.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.7 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 0.7 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.2 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 3.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 0.9 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.2 | 2.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 5.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 2.9 | GO:0005624 | obsolete membrane fraction(GO:0005624) |
| 0.1 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 3.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.7 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.1 | 3.0 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.2 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 1.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.6 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.9 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 5.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 1.1 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 2.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 2.3 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.0 | 0.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.9 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0010008 | endosome membrane(GO:0010008) endosomal part(GO:0044440) |
| 0.0 | 12.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 2.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 2.5 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 0.6 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.4 | 5.5 | GO:0008907 | integrase activity(GO:0008907) |
| 0.6 | 2.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.6 | 4.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.6 | 1.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.5 | 1.5 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-2 binding(GO:0019976) |
| 0.5 | 3.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.4 | 6.0 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.4 | 5.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.4 | 1.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 1.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.7 | GO:0004803 | transposase activity(GO:0004803) |
| 0.3 | 1.0 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.3 | 1.6 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.3 | 3.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 0.9 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.3 | 1.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.3 | 2.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 0.9 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.3 | 0.8 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.3 | 2.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.3 | 2.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.3 | 0.8 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.3 | 2.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 0.7 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.2 | 3.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 1.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 1.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 4.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 0.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 0.6 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.2 | 1.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 0.6 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 5.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 2.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 2.8 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.5 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 3.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 0.9 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.2 | 0.5 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 1.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 0.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 1.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 1.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 3.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 0.4 | GO:0031997 | N-terminal myristoylation domain binding(GO:0031997) |
| 0.1 | 0.4 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 1.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 3.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.8 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.1 | 0.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.5 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.3 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.4 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 2.7 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 1.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.7 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.5 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 1.8 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.6 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.5 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.1 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.3 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.1 | 0.6 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 0.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.4 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.1 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 1.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.9 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 0.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.1 | 0.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 3.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 6.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.5 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 3.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.1 | GO:0047042 | androsterone dehydrogenase (B-specific) activity(GO:0047042) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 3.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.4 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 6.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.6 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 4.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 1.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.0 | GO:0003704 | obsolete specific RNA polymerase II transcription factor activity(GO:0003704) |
| 0.0 | 0.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.4 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 2.3 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 2.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 9.5 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 4.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.3 | GO:0030528 | obsolete transcription regulator activity(GO:0030528) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 3.6 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 1.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.7 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.0 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.0 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.5 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.2 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 0.4 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 6.4 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.4 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.1 | 0.8 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 2.4 | ST_B_CELL_ANTIGEN_RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 1.5 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 0.6 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.1 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.7 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.1 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.2 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |