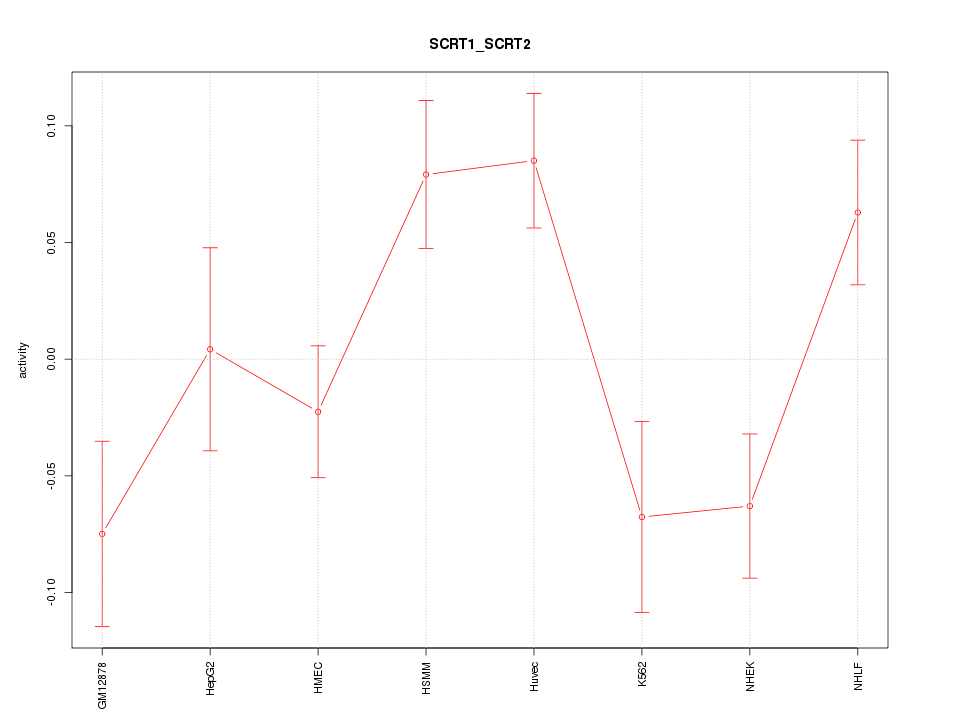
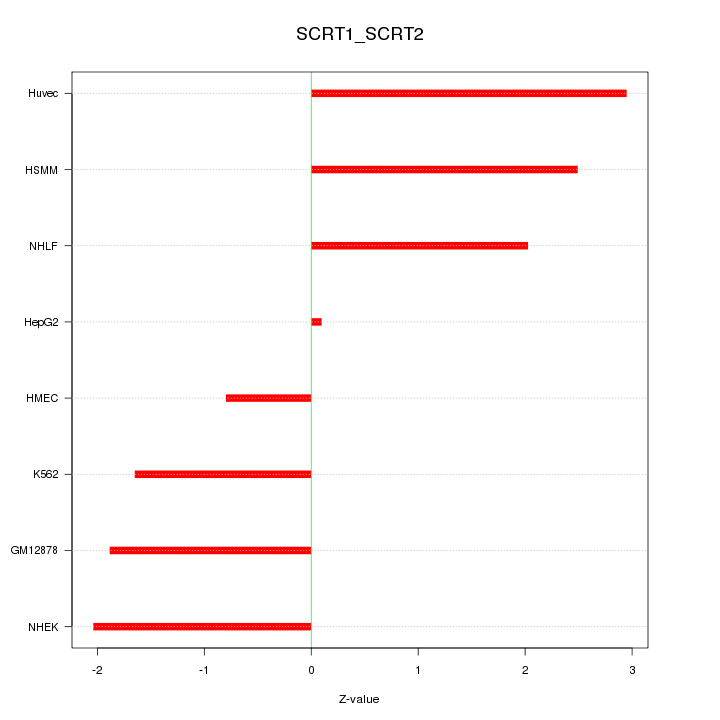
Motif ID: SCRT1_SCRT2
Z-value: 1.940
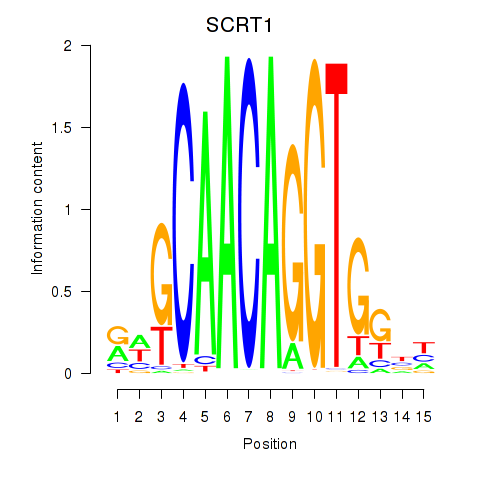
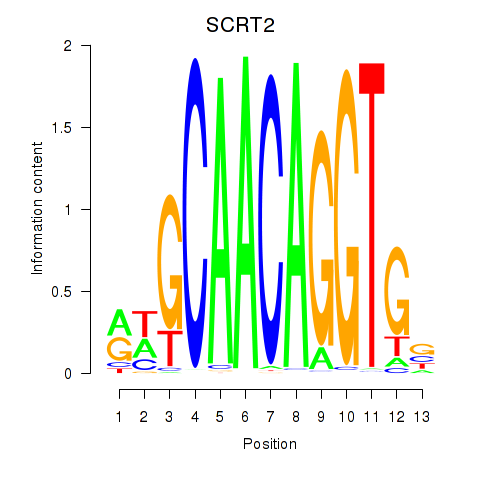
Transcription factors associated with SCRT1_SCRT2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SCRT1 | ENSG00000170616.9 | SCRT1 |
| SCRT2 | ENSG00000215397.3 | SCRT2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.3 | 5.3 | GO:0051503 | purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 0.8 | 2.5 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.5 | 7.5 | GO:0000052 | citrulline metabolic process(GO:0000052) arginine catabolic process(GO:0006527) |
| 0.3 | 1.3 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.3 | 0.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 0.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 2.7 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.2 | 0.8 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 0.9 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 | 0.4 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.3 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 1.1 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.1 | 0.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.8 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 3.8 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.1 | 0.2 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.3 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.1 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 4.6 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 4.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:0071230 | positive regulation of TOR signaling(GO:0032008) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.2 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.2 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.7 | GO:0036503 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.5 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 1.1 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.5 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 1.4 | GO:0034339 | obsolete regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor(GO:0034339) |
| 0.0 | 0.2 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.0 | 0.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.7 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 4.3 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.3 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 | 0.8 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.7 | GO:0007623 | circadian rhythm(GO:0007623) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 0.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 2.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.8 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 3.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 6.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 2.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) histone pre-mRNA 3'end processing complex(GO:0071204) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 1.2 | 6.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 5.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 3.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 1.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 2.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.2 | GO:0019862 | calcium channel inhibitor activity(GO:0019855) IgA binding(GO:0019862) |
| 0.1 | 0.3 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 1.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 4.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.5 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.6 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.1 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |