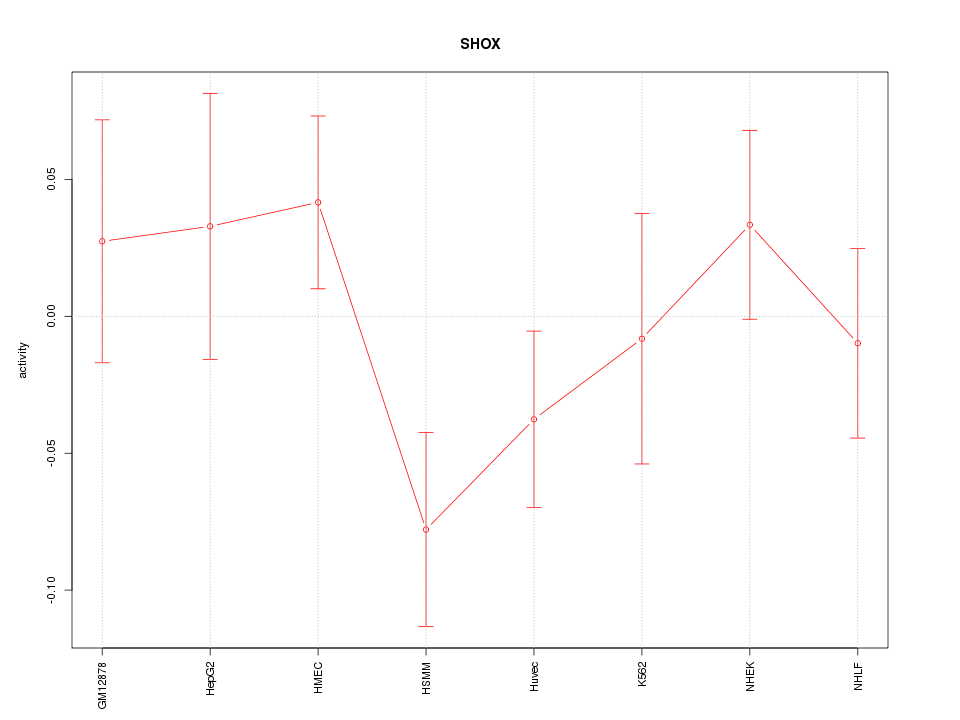
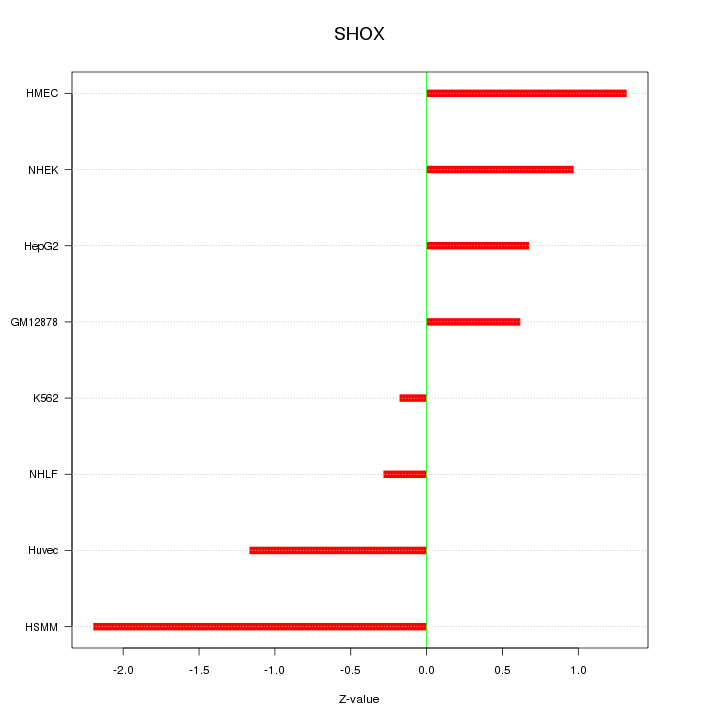
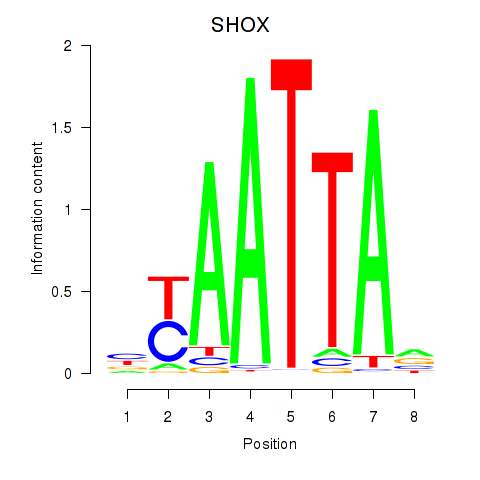
Motif ID: SHOX
Z-value: 1.108
Transcription factors associated with SHOX:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SHOX | ENSG00000185960.8 | SHOX |
| SHOX | ENSGR0000185960.8 | SHOX |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0060649 | nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.4 | 1.6 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.4 | 1.1 | GO:0035987 | endodermal cell fate commitment(GO:0001711) endodermal cell differentiation(GO:0035987) |
| 0.4 | 1.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.3 | 0.9 | GO:0055005 | optic placode formation involved in camera-type eye formation(GO:0046619) ventricular cardiac myofibril assembly(GO:0055005) regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.2 | 0.7 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 1.3 | GO:0043316 | natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.4 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 | 0.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.1 | 0.6 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.4 | GO:0042097 | lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.5 | GO:0046621 | negative regulation of organ growth(GO:0046621) |
| 0.1 | 0.3 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.3 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.1 | 0.2 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 0.7 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.6 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.6 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.1 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.1 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.2 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.3 | GO:0032472 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.8 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.3 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) menarche(GO:0042696) |
| 0.0 | 1.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0045938 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.5 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.4 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.7 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) skeletal muscle organ development(GO:0060538) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0035444 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) ferrous iron transport(GO:0015684) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) ferrous iron import(GO:0070627) iron ion import(GO:0097286) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.7 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.2 | GO:0060042 | retina morphogenesis in camera-type eye(GO:0060042) |
| 0.0 | 0.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.9 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.1 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 0.2 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.3 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 3.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) synaptic cleft(GO:0043083) |
| 0.1 | 0.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0036452 | ESCRT III complex(GO:0000815) ESCRT complex(GO:0036452) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.0 | GO:0032584 | growth cone membrane(GO:0032584) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 0.9 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 1.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.1 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.1 | 0.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.9 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 2.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.4 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.5 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) SAM domain binding(GO:0032093) |
| 0.1 | 1.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.3 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 6.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) ferrous iron transmembrane transporter activity(GO:0015093) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) chromium ion transmembrane transporter activity(GO:0070835) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.8 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | ST_B_CELL_ANTIGEN_RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |