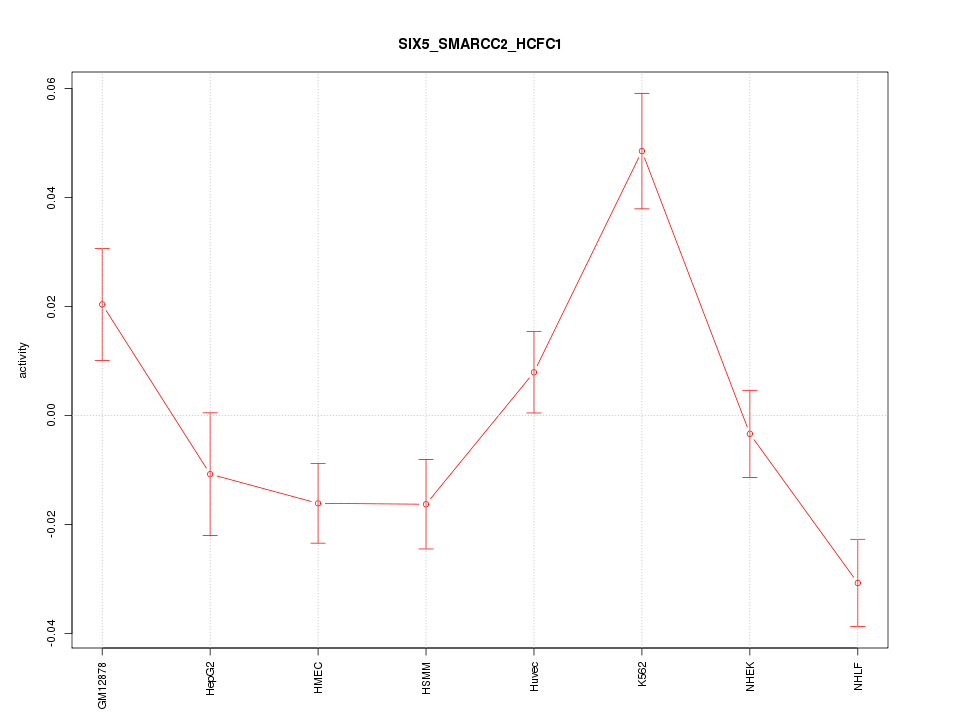
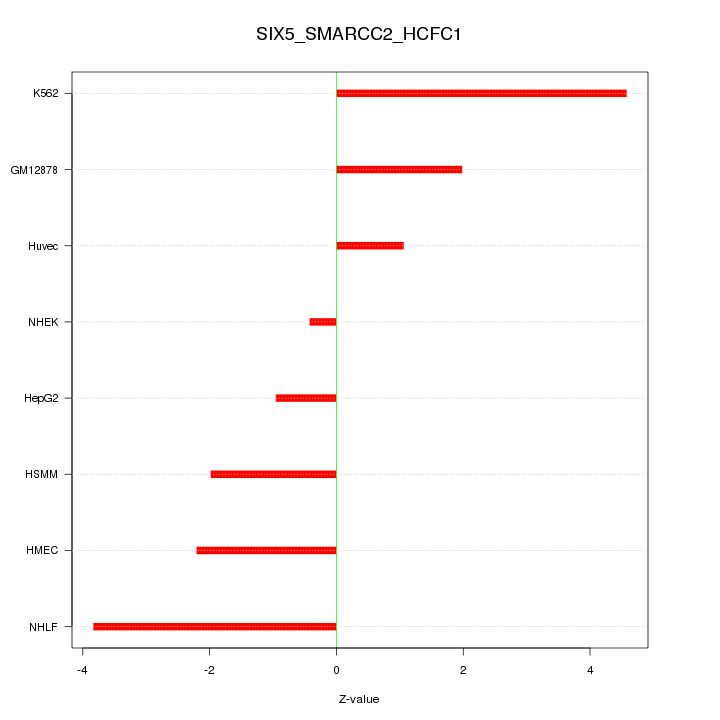
Motif ID: SIX5_SMARCC2_HCFC1
Z-value: 2.515
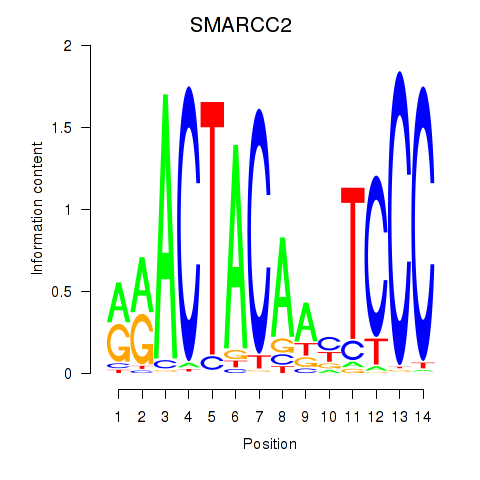
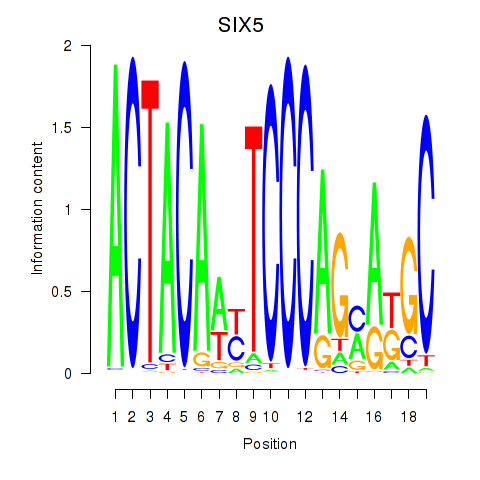
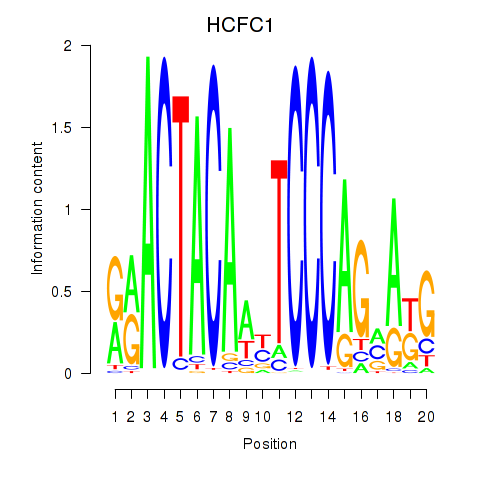
Transcription factors associated with SIX5_SMARCC2_HCFC1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HCFC1 | ENSG00000172534.9 | HCFC1 |
| SIX5 | ENSG00000177045.6 | SIX5 |
| SMARCC2 | ENSG00000139613.7 | SMARCC2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.7 | 7.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.7 | 2.7 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.5 | 2.1 | GO:0070922 | small RNA loading onto RISC(GO:0070922) |
| 0.5 | 2.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 1.3 | GO:0032196 | transposition, DNA-mediated(GO:0006313) transposition(GO:0032196) |
| 0.4 | 1.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 | 1.1 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.3 | 1.0 | GO:0015854 | guanine transport(GO:0015854) hypoxanthine transport(GO:0035344) thymine transport(GO:0035364) |
| 0.3 | 1.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.3 | 1.0 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) protein hexamerization(GO:0034214) |
| 0.3 | 0.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.3 | 2.1 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.3 | 0.9 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.3 | 0.9 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.3 | 0.9 | GO:0046368 | GDP-L-fucose biosynthetic process(GO:0042350) 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) GDP-L-fucose metabolic process(GO:0046368) |
| 0.3 | 1.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 1.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 1.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.3 | 1.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.3 | 0.8 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.3 | 1.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.3 | 0.3 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.2 | 1.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 1.0 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.2 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 0.9 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.2 | 1.8 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.2 | 1.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.2 | 0.6 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.2 | 1.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.2 | 2.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 1.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.7 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 0.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 1.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.2 | 0.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.2 | 0.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.5 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.1 | 0.9 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 1.4 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 1.8 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 1.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.7 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.8 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 1.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.4 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 1.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.4 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.1 | 0.7 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 2.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.1 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.1 | 1.9 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.1 | 1.0 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.1 | 0.4 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.6 | GO:0032057 | negative regulation of translation in response to stress(GO:0032055) negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 2.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.9 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.5 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.9 | GO:0098534 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.1 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.3 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.1 | 0.3 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.5 | GO:0032527 | retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.1 | 0.3 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.9 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.2 | GO:0002431 | Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 1.8 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.7 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.1 | 0.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 3.7 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.1 | 1.2 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 1.8 | GO:0044349 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.1 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.1 | 0.9 | GO:0009220 | pyrimidine ribonucleotide biosynthetic process(GO:0009220) |
| 0.1 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.2 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.1 | 0.9 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.4 | GO:0061339 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0031167 | rRNA modification(GO:0000154) rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.7 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.9 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 1.8 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 6.2 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 1.4 | GO:0045892 | negative regulation of cellular biosynthetic process(GO:0031327) negative regulation of transcription, DNA-templated(GO:0045892) negative regulation of RNA metabolic process(GO:0051253) negative regulation of RNA biosynthetic process(GO:1902679) negative regulation of nucleic acid-templated transcription(GO:1903507) |
| 0.0 | 0.4 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 2.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.9 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.1 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.3 | GO:0060119 | auditory receptor cell development(GO:0060117) inner ear receptor cell development(GO:0060119) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 3.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0046039 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) GTP metabolic process(GO:0046039) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0071474 | cellular water homeostasis(GO:0009992) cellular hyperosmotic response(GO:0071474) |
| 0.0 | 1.5 | GO:0001892 | embryonic placenta development(GO:0001892) |
| 0.0 | 0.1 | GO:0060004 | semicircular canal morphogenesis(GO:0048752) reflex(GO:0060004) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.4 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.5 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 2.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of monooxygenase activity(GO:0032769) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0072207 | metanephric tubule development(GO:0072170) metanephric epithelium development(GO:0072207) metanephric nephron tubule development(GO:0072234) metanephric nephron epithelium development(GO:0072243) |
| 0.0 | 1.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0044060 | regulation of endocrine process(GO:0044060) |
| 0.0 | 2.4 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.5 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.4 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.3 | GO:0045069 | regulation of viral genome replication(GO:0045069) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.7 | GO:0016568 | chromatin modification(GO:0016568) |
| 0.0 | 0.2 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0052646 | phosphatidylserine metabolic process(GO:0006658) alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.2 | GO:0070536 | G2 DNA damage checkpoint(GO:0031572) protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.7 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.7 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.1 | GO:0098657 | neurotransmitter uptake(GO:0001504) import into cell(GO:0098657) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.4 | GO:0050931 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 | 0.1 | GO:0007005 | mitochondrion organization(GO:0007005) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0000018 | regulation of DNA recombination(GO:0000018) |
| 0.0 | 0.3 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.0 | 0.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.0 | GO:0052257 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.7 | 7.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.5 | 2.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 1.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 2.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.4 | 1.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.3 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 0.6 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.3 | 0.8 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.3 | 1.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 0.8 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 2.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 0.9 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 1.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 1.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 1.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.3 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) endoribonuclease complex(GO:1902555) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 2.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.8 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 1.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.7 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.7 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.1 | 1.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.7 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 1.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.6 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 2.6 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.3 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.1 | 0.2 | GO:0005652 | lamin filament(GO:0005638) nuclear lamina(GO:0005652) |
| 0.0 | 0.6 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.8 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 2.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 6.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0044432 | endoplasmic reticulum part(GO:0044432) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.0 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.6 | GO:0044448 | cell cortex part(GO:0044448) |
| 0.0 | 0.3 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 14.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.6 | 2.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.6 | 2.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.5 | 1.5 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.5 | 2.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.4 | 1.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.4 | 2.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.4 | 1.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.3 | 2.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 1.3 | GO:0004803 | transposase activity(GO:0004803) |
| 0.3 | 1.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 0.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 1.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.9 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.2 | 1.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.2 | 0.6 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.2 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 0.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 0.7 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.2 | 3.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 1.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.2 | 0.9 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 1.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 2.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 0.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.4 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.1 | 1.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.8 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.4 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 0.5 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 1.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.6 | GO:0008753 | NADPH dehydrogenase (quinone) activity(GO:0008753) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 2.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.1 | 0.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.4 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.4 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 1.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 3.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.7 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 3.6 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.6 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 1.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.0 | 0.9 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.0 | 1.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 3.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 4.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.6 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0016563 | obsolete transcription activator activity(GO:0016563) |
| 0.0 | 2.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 3.8 | GO:0010843 | obsolete promoter binding(GO:0010843) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 2.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 1.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0008907 | integrase activity(GO:0008907) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0016566 | obsolete specific transcriptional repressor activity(GO:0016566) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 1.5 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.1 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 2.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.7 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |