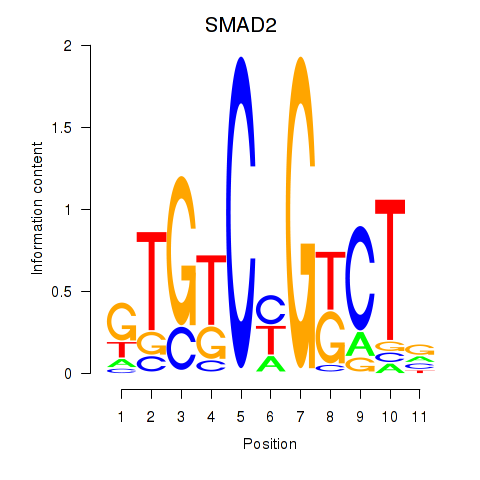
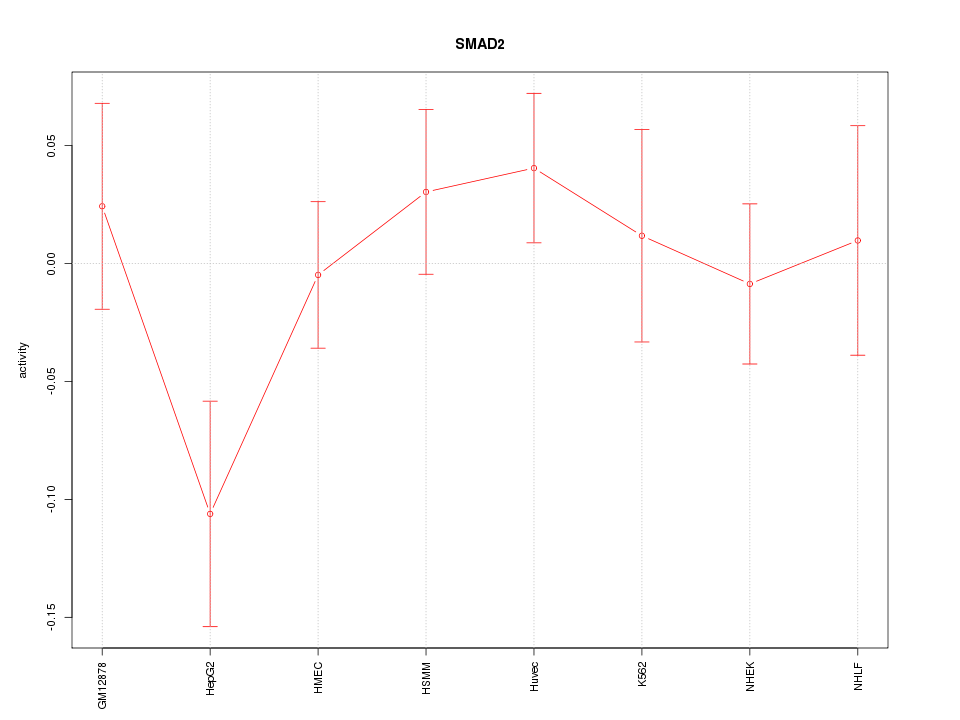
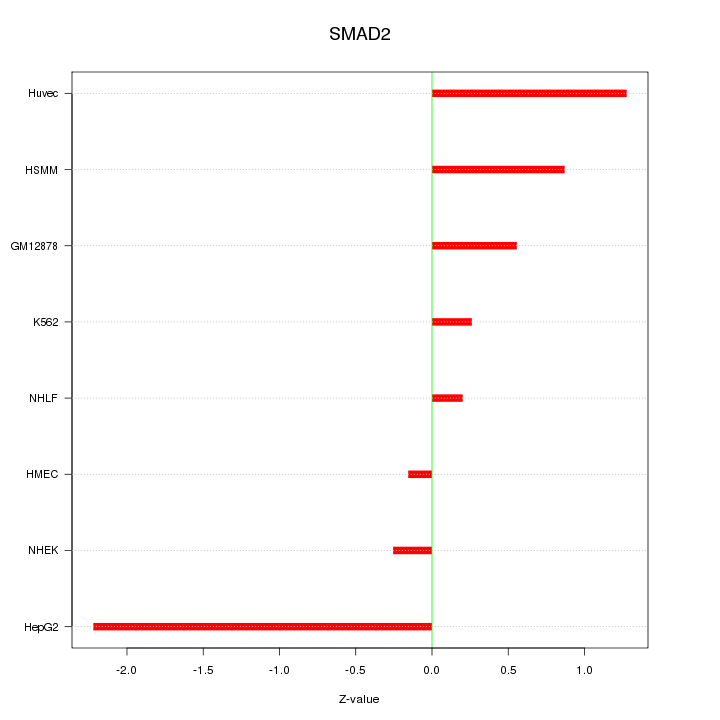
| 0.3 |
1.0 |
GO:0021570 |
rhombomere 4 development(GO:0021570) |
| 0.2 |
0.9 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 |
0.7 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.1 |
0.4 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 |
1.1 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 |
0.5 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.1 |
0.5 |
GO:2000008 |
regulation of protein localization to cell surface(GO:2000008) |
| 0.1 |
3.5 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
0.6 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.3 |
GO:0014724 |
regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) mitochondrial calcium ion homeostasis(GO:0051560) positive regulation of mitochondrial calcium ion concentration(GO:0051561) relaxation of skeletal muscle(GO:0090076) |
| 0.1 |
0.4 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.1 |
0.6 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.1 |
1.2 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.1 |
0.5 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
0.6 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
0.4 |
GO:0006216 |
cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 |
0.6 |
GO:0045010 |
actin nucleation(GO:0045010) |
| 0.1 |
1.3 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 |
1.2 |
GO:0045749 |
obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.1 |
0.3 |
GO:0045046 |
protein import into peroxisome membrane(GO:0045046) |
| 0.1 |
0.4 |
GO:0090281 |
negative regulation of neurotransmitter transport(GO:0051589) negative regulation of calcium ion import(GO:0090281) |
| 0.1 |
0.3 |
GO:0045988 |
negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 |
0.5 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.1 |
GO:0097709 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 |
0.1 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.0 |
0.2 |
GO:0032236 |
obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 |
0.1 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
1.0 |
GO:0042116 |
macrophage activation(GO:0042116) |
| 0.0 |
0.6 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.2 |
GO:0034204 |
lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 |
0.7 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 |
0.5 |
GO:0046135 |
pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.1 |
GO:0032928 |
regulation of superoxide anion generation(GO:0032928) negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.1 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 |
0.2 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 |
0.1 |
GO:0070781 |
response to biotin(GO:0070781) |
| 0.0 |
0.2 |
GO:1903078 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 |
0.7 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.5 |
GO:0006513 |
protein monoubiquitination(GO:0006513) |
| 0.0 |
0.3 |
GO:0006656 |
phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 |
0.1 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.5 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.2 |
GO:0045987 |
positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 |
0.5 |
GO:0016525 |
negative regulation of angiogenesis(GO:0016525) |
| 0.0 |
1.7 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.0 |
0.0 |
GO:0008300 |
isoprenoid catabolic process(GO:0008300) |
| 0.0 |
0.1 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.0 |
GO:0019056 |
modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 |
0.1 |
GO:0046618 |
drug transmembrane transport(GO:0006855) tetracycline transport(GO:0015904) drug export(GO:0046618) |
| 0.0 |
0.4 |
GO:0000080 |
mitotic G1 phase(GO:0000080) |
| 0.0 |
0.0 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
1.0 |
GO:0008360 |
regulation of cell shape(GO:0008360) |