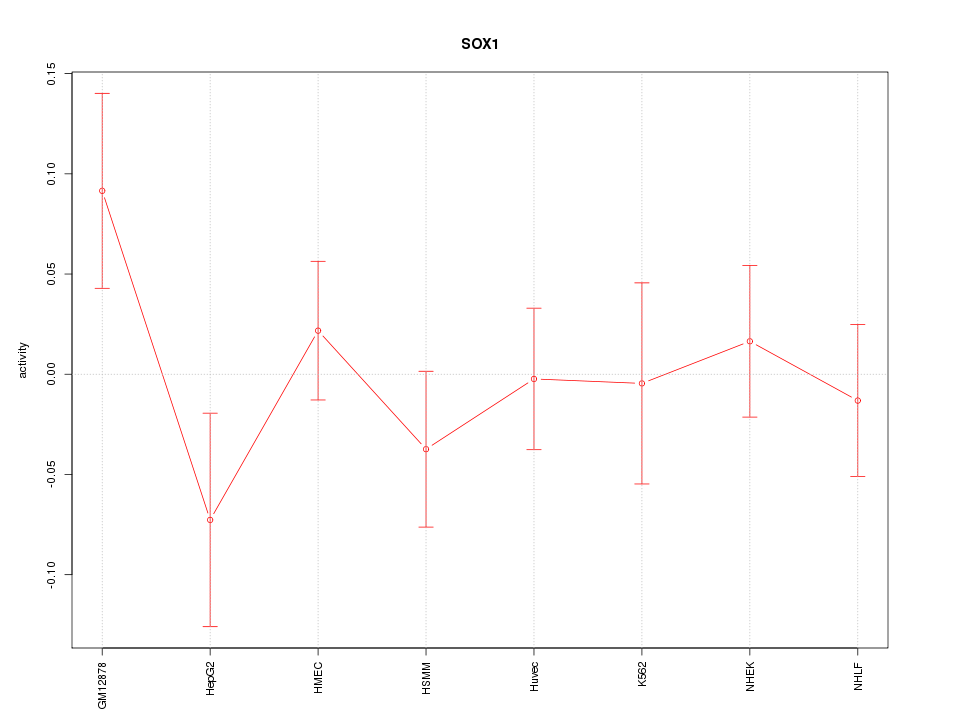
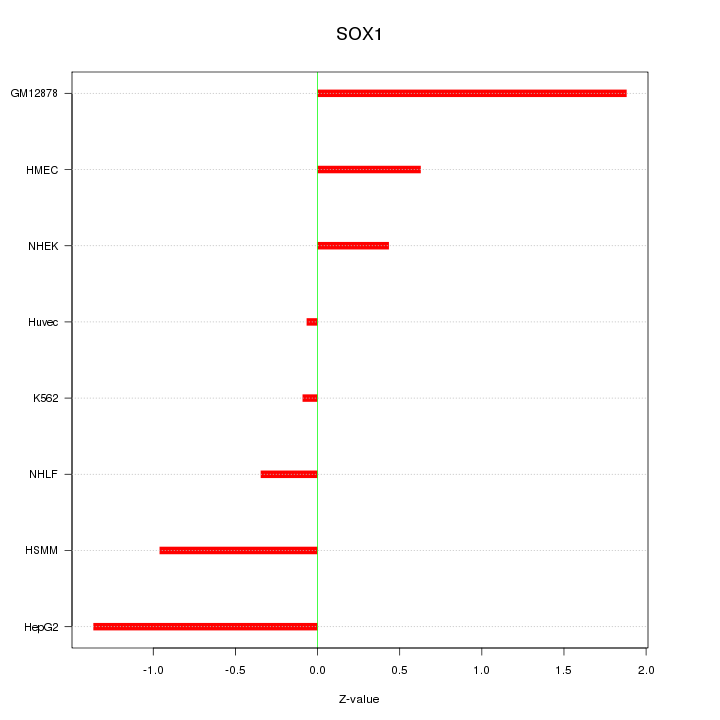
Motif ID: SOX1
Z-value: 0.938
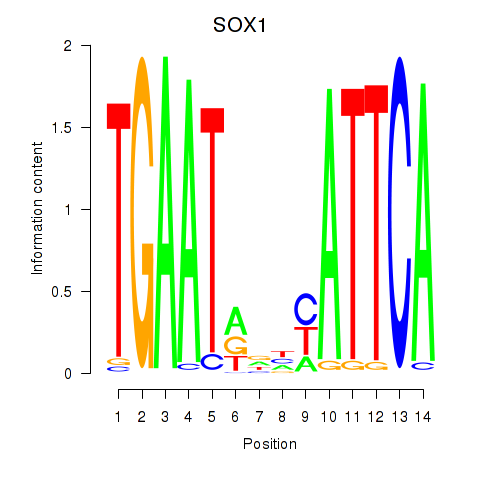
Transcription factors associated with SOX1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SOX1 | ENSG00000182968.3 | SOX1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.4 | 10.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.3 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.1 | 0.2 | GO:0070340 | cell surface pattern recognition receptor signaling pathway(GO:0002752) detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.6 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.3 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.0 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0060122 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.1 | GO:1901160 | serotonin metabolic process(GO:0042428) primary amino compound metabolic process(GO:1901160) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.6 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 2.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.6 | 10.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.7 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.8 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) lipopeptide binding(GO:0071723) |
| 0.1 | 1.0 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.0 | GO:0016297 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.6 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |