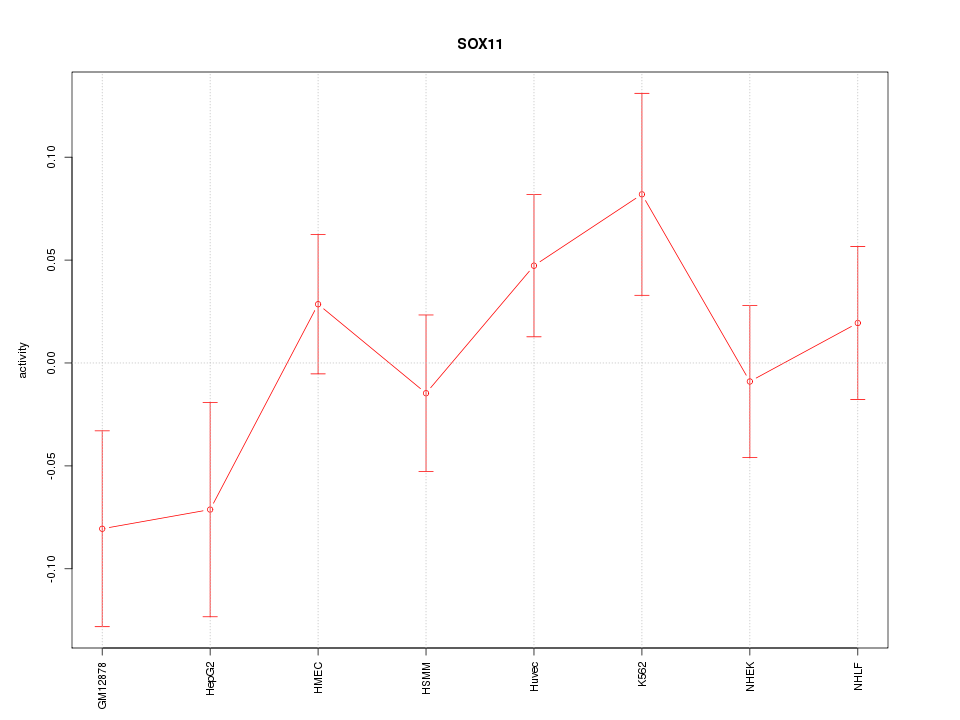
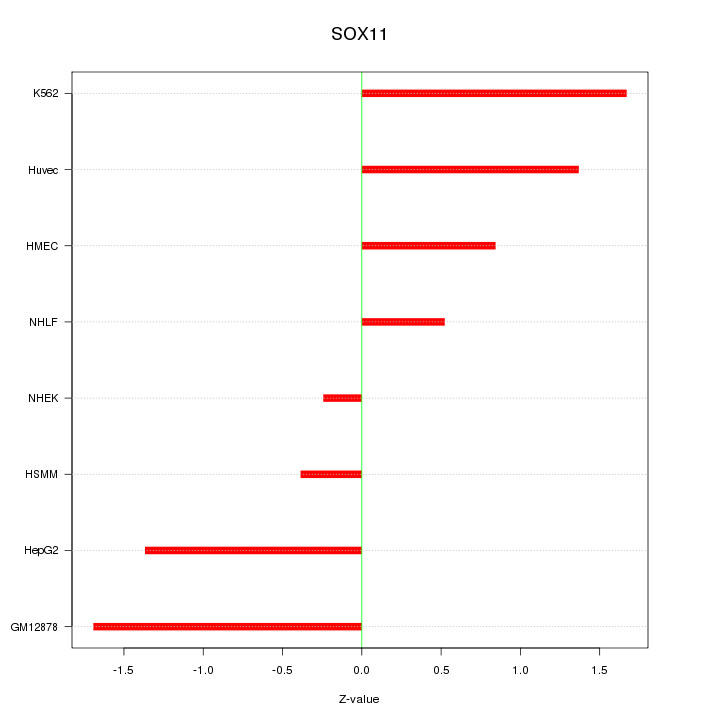
Motif ID: SOX11
Z-value: 1.151
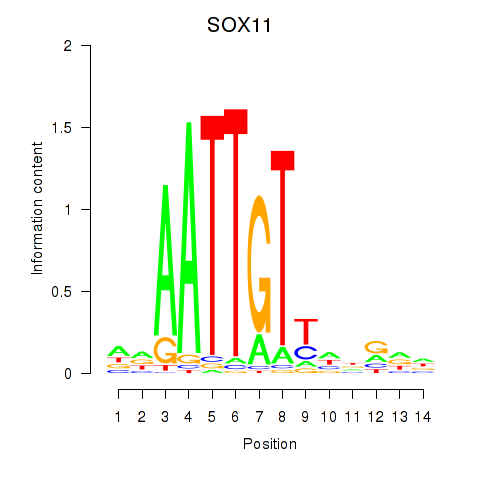
Transcription factors associated with SOX11:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SOX11 | ENSG00000176887.5 | SOX11 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 | 2.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 1.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 0.9 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.2 | 1.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.6 | GO:0034371 | positive regulation of sequestering of triglyceride(GO:0010890) chylomicron remodeling(GO:0034371) |
| 0.1 | 1.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.1 | 2.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.3 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) small RNA loading onto RISC(GO:0070922) |
| 0.1 | 0.5 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 0.5 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 | 0.6 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0071418 | uroporphyrinogen III biosynthetic process(GO:0006780) cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 1.1 | GO:0050931 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 | 0.2 | GO:0060122 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0050912 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 2.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 3.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.1 | GO:0098858 | actin-based cell projection(GO:0098858) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 0.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 1.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 1.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 0.6 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 1.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 0.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 0.5 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.4 | GO:0050833 | pyruvate secondary active transmembrane transporter activity(GO:0005477) pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.7 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |