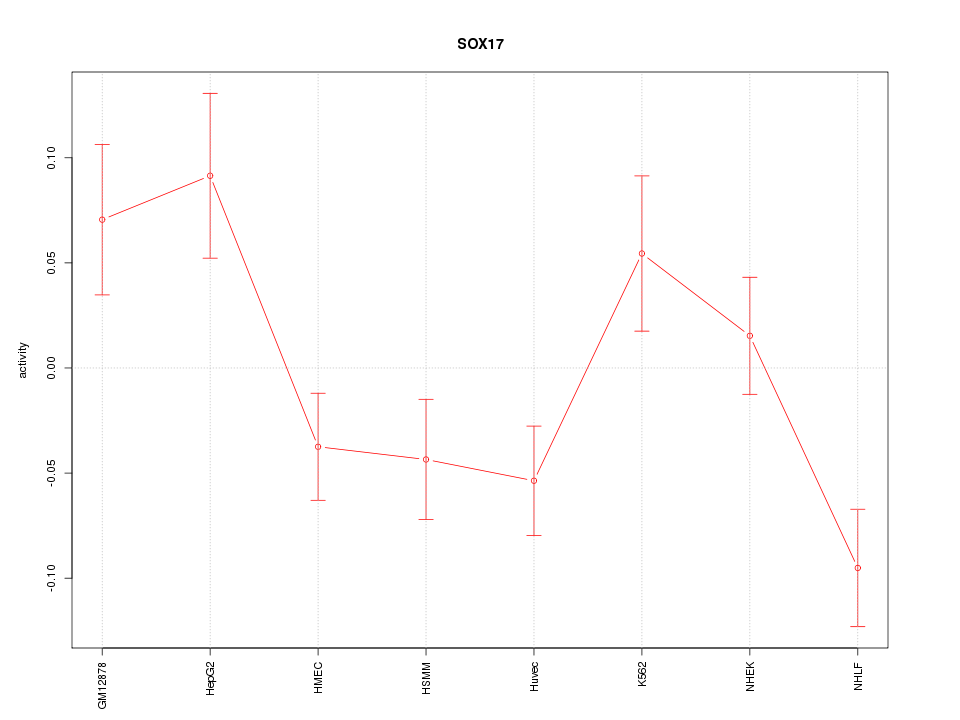
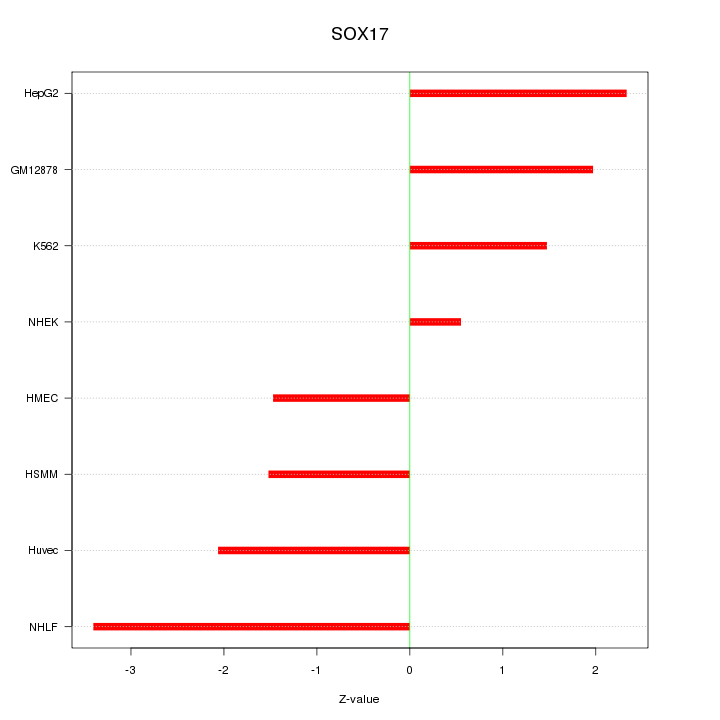
Motif ID: SOX17
Z-value: 2.004
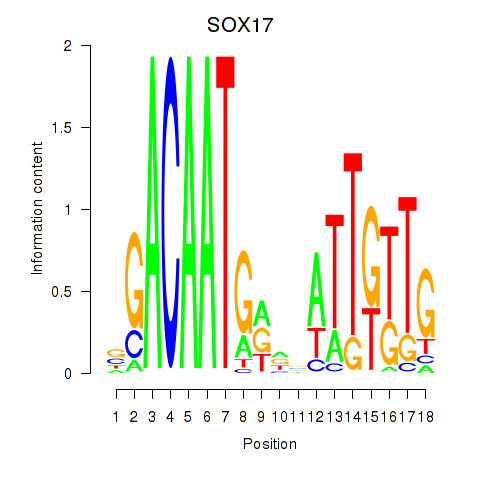
Transcription factors associated with SOX17:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SOX17 | ENSG00000164736.5 | SOX17 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0010903 | negative regulation of cytokine secretion involved in immune response(GO:0002740) negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.7 | 2.0 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.6 | 1.9 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.4 | 1.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.4 | 1.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.4 | 1.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.4 | 1.4 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.3 | 2.7 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.3 | 2.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 | 1.3 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 1.2 | GO:0042905 | retinal metabolic process(GO:0042574) 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 2.9 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.2 | 1.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 0.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 3.1 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.5 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.7 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.1 | 0.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.6 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.6 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.2 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.3 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.1 | 0.2 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.1 | 1.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.2 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.1 | 0.7 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 0.4 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.6 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 1.8 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.3 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 2.0 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 1.2 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 1.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.5 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 1.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.3 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.5 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 2.3 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.7 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.8 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 | 6.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.2 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.5 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 1.3 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) negative regulation of oligodendrocyte differentiation(GO:0048715) positive regulation of dopamine receptor signaling pathway(GO:0060161) negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 1.2 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.2 | GO:0050704 | interleukin-1 beta secretion(GO:0050702) regulation of interleukin-1 secretion(GO:0050704) regulation of interleukin-1 beta secretion(GO:0050706) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.3 | 3.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.6 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 5.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.2 | 0.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 3.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.1 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.5 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 3.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.5 | 2.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 2.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.4 | 2.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 2.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.3 | 1.9 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.3 | 2.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 0.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 1.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 1.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.6 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 1.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 1.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.3 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 1.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 2.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 2.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 3.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.4 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 2.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 2.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 1.4 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 2.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |