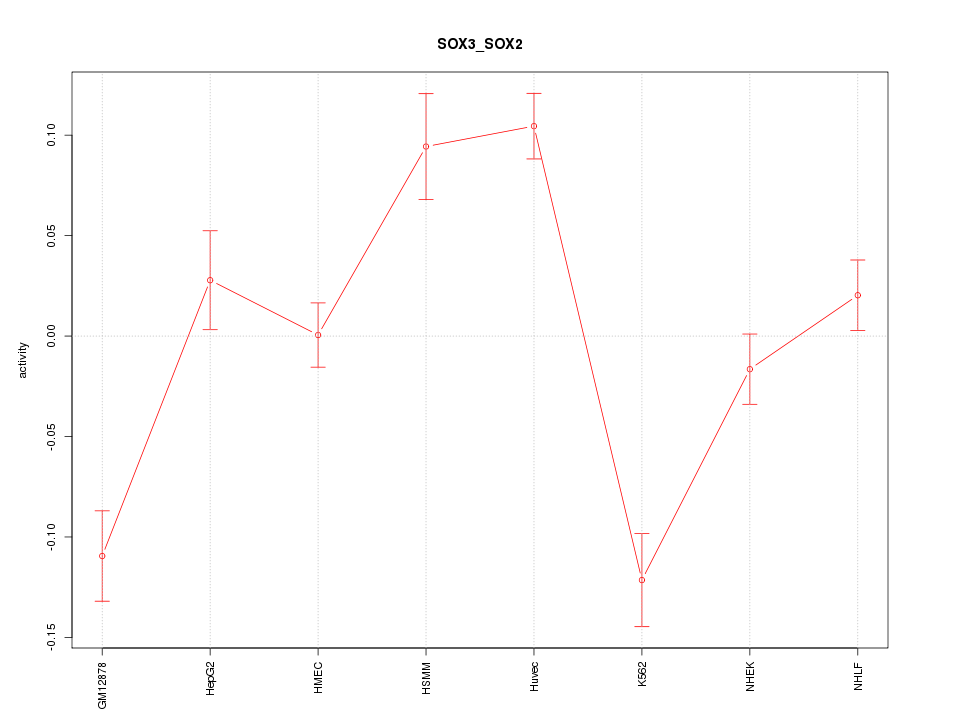
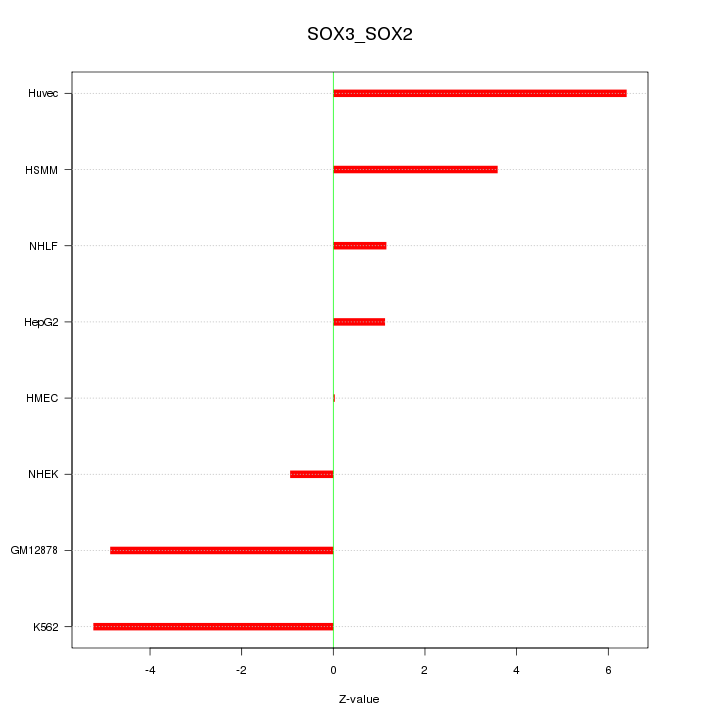
Motif ID: SOX3_SOX2
Z-value: 3.682
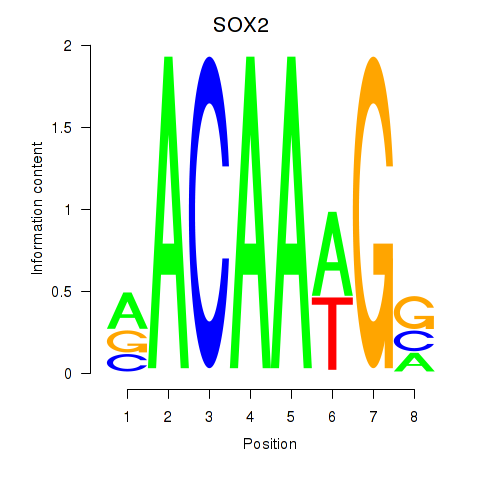
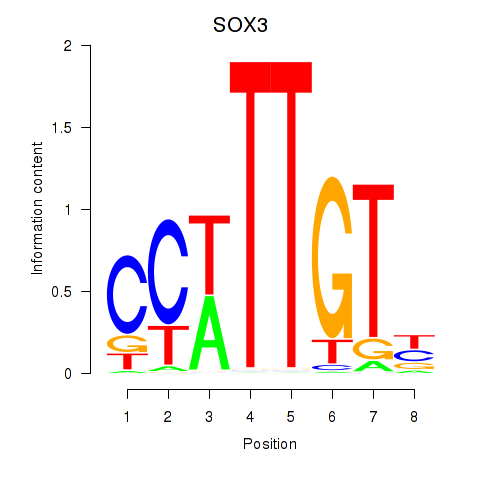
Transcription factors associated with SOX3_SOX2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SOX2 | ENSG00000181449.2 | SOX2 |
| SOX3 | ENSG00000134595.6 | SOX3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 3.2 | 9.7 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 1.6 | 4.9 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 1.5 | 9.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.2 | 3.5 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) regulation of mesodermal cell fate specification(GO:0042661) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of mesoderm development(GO:2000380) |
| 1.0 | 2.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 1.0 | 4.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.8 | 2.5 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.8 | 5.8 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 | 5.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.8 | 5.0 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.8 | 3.3 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.8 | 2.4 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.8 | 11.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.7 | 0.7 | GO:0051893 | regulation of focal adhesion assembly(GO:0051893) negative regulation of focal adhesion assembly(GO:0051895) regulation of cell-substrate junction assembly(GO:0090109) regulation of cell junction assembly(GO:1901888) negative regulation of cell junction assembly(GO:1901889) regulation of adherens junction organization(GO:1903391) negative regulation of adherens junction organization(GO:1903392) |
| 0.7 | 9.5 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.6 | 3.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.5 | 1.0 | GO:0070255 | regulation of mucus secretion(GO:0070255) negative regulation of mucus secretion(GO:0070256) |
| 0.5 | 5.5 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.5 | 1.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.5 | 2.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.5 | 1.5 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.5 | 1.9 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.5 | 1.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 1.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 | 3.4 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.4 | 2.5 | GO:0099515 | nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.4 | 2.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.4 | 2.8 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.4 | 3.5 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.4 | 4.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.4 | 9.5 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.4 | 1.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.3 | 3.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.3 | 1.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.3 | 1.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.3 | 1.0 | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.3 | 2.7 | GO:0003065 | regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.3 | 0.3 | GO:0021873 | forebrain neuroblast division(GO:0021873) neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.3 | 2.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 5.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 7.9 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.3 | 1.3 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.3 | 1.6 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.3 | 1.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 | 1.2 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.2 | 0.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.2 | 0.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 0.4 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.2 | 0.7 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.2 | 0.6 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.2 | 1.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 0.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 3.1 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 4.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 1.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 0.6 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.2 | 8.0 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.2 | 1.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.9 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 | 0.7 | GO:0034959 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.2 | 1.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.2 | 1.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.2 | 1.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 0.3 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.2 | 3.0 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.2 | 5.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 2.2 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.2 | 13.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.2 | 1.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 0.3 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 1.3 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.2 | 1.2 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.2 | 3.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.2 | 2.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 0.6 | GO:0072367 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.8 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 1.9 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 2.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 2.7 | GO:0055007 | cardiac muscle cell differentiation(GO:0055007) |
| 0.1 | 0.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.7 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.1 | 2.0 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.1 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.4 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.8 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.5 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 1.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 2.2 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.1 | 0.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 3.8 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 0.3 | GO:0021604 | facial nerve development(GO:0021561) rhombomere 4 development(GO:0021570) cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) |
| 0.1 | 3.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 0.5 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 3.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 6.8 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.1 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.4 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.1 | 1.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.7 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 0.3 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.1 | 0.3 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.6 | GO:0046349 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 3.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.2 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 3.3 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.3 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.1 | 1.4 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 1.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.1 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.5 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 0.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 0.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 8.3 | GO:0008154 | actin polymerization or depolymerization(GO:0008154) |
| 0.1 | 0.6 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.1 | 0.7 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 1.2 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.1 | GO:0060346 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.1 | 0.7 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 0.4 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 | 0.8 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 4.4 | GO:0070252 | actin-mediated cell contraction(GO:0070252) |
| 0.1 | 0.5 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.5 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.1 | 0.2 | GO:0032196 | transposition, DNA-mediated(GO:0006313) transposition(GO:0032196) |
| 0.1 | 0.5 | GO:0030277 | epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.0 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 0.8 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.1 | 0.2 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 1.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 5.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 2.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 4.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 1.2 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 1.8 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 0.4 | GO:0060008 | Sertoli cell differentiation(GO:0060008) Sertoli cell development(GO:0060009) |
| 0.1 | 12.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 8.1 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 0.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.3 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.9 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.1 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.6 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.2 | GO:0030163 | protein catabolic process(GO:0030163) |
| 0.0 | 0.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:1903301 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 5.7 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 6.9 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.8 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.1 | GO:0071801 | podosome assembly(GO:0071800) regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 4.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0006547 | histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.0 | 1.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 1.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0009071 | glycine metabolic process(GO:0006544) glycine catabolic process(GO:0006546) serine family amino acid catabolic process(GO:0009071) |
| 0.0 | 0.7 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 9.3 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.4 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 5.8 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 1.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 7.1 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.7 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0045768 | obsolete positive regulation of anti-apoptosis(GO:0045768) |
| 0.0 | 0.1 | GO:0002237 | response to molecule of bacterial origin(GO:0002237) response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 1.8 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.5 | GO:0034764 | positive regulation of transmembrane transport(GO:0034764) positive regulation of ion transmembrane transport(GO:0034767) |
| 0.0 | 0.1 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 4.7 | GO:0071466 | xenobiotic metabolic process(GO:0006805) response to xenobiotic stimulus(GO:0009410) cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 2.0 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.3 | GO:0072164 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.9 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:0009187 | cyclic nucleotide metabolic process(GO:0009187) |
| 0.0 | 0.5 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.5 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 1.1 | GO:0071772 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.1 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.4 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.2 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.3 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 1.6 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 1.1 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.0 | 2.0 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.0 | 0.8 | GO:0051297 | microtubule organizing center organization(GO:0031023) centrosome organization(GO:0051297) |
| 0.0 | 1.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.7 | GO:0051258 | protein polymerization(GO:0051258) |
| 0.0 | 0.1 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.3 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.3 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0031670 | cellular response to nutrient(GO:0031670) |
| 0.0 | 0.4 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.1 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:0051402 | neuron apoptotic process(GO:0051402) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 15.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 1.1 | 5.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.0 | 3.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.6 | 5.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.6 | 9.7 | GO:0001527 | microfibril(GO:0001527) |
| 0.5 | 3.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.4 | 2.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 3.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 2.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 2.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 4.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 2.8 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 5.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 0.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.3 | 3.4 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 3.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.3 | 2.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 4.5 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 1.7 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.2 | 11.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 1.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 3.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 8.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 1.0 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.2 | 2.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 16.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 47.5 | GO:0005912 | adherens junction(GO:0005912) |
| 0.2 | 1.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 12.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.2 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 13.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 3.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 1.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.1 | 3.1 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.5 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 0.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 13.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.2 | GO:0043205 | fibril(GO:0043205) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.6 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.1 | 2.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.6 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.1 | 1.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 1.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.1 | 2.4 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.1 | 5.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 22.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.2 | GO:0036452 | ESCRT III complex(GO:0000815) ESCRT complex(GO:0036452) |
| 0.1 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 33.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.8 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.6 | GO:0030684 | preribosome(GO:0030684) |
| 0.0 | 4.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.8 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.0 | 5.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 4.0 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 9.8 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.1 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 4.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.0 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.2 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.8 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 5.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) axoneme part(GO:0044447) |
| 0.0 | 1.2 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 3.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.5 | 6.2 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 1.3 | 12.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.2 | 5.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.0 | 11.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.9 | 9.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.9 | 7.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.8 | 3.4 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.8 | 2.4 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.8 | 3.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.6 | 9.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.6 | 1.9 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.5 | 12.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 8.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 2.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 2.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.4 | 4.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.4 | 8.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 1.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.4 | 1.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 1.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.3 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 3.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 5.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 8.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.3 | 0.9 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.3 | 1.5 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 1.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.3 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.3 | 0.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 1.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.3 | 1.4 | GO:0004741 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 5.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 10.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 1.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) proteoglycan sulfotransferase activity(GO:0050698) |
| 0.3 | 3.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 0.8 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.2 | 1.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 1.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 12.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 1.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 0.9 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.2 | 1.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 3.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.6 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 0.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 5.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 0.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 1.6 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 0.9 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.2 | 15.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 2.6 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.2 | 7.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 0.6 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.2 | 2.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 3.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.1 | 1.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 1.2 | GO:0051425 | acetylcholine receptor binding(GO:0033130) PTB domain binding(GO:0051425) |
| 0.1 | 2.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 1.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 2.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 4.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.9 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 4.6 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 3.0 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.5 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.4 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.8 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.3 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.6 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 12.8 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 2.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.6 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 2.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.6 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 15.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 9.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.9 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 5.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 1.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 6.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 1.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 1.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) poly-purine tract binding(GO:0070717) |
| 0.1 | 2.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.5 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 11.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 1.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 1.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 7.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 12.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 11.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.9 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 3.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0016566 | obsolete specific transcriptional repressor activity(GO:0016566) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 9.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.9 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 1.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 10.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.0 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 1.3 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 0.4 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.2 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.3 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 1.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.0 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.2 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.7 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.8 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.5 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.1 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |