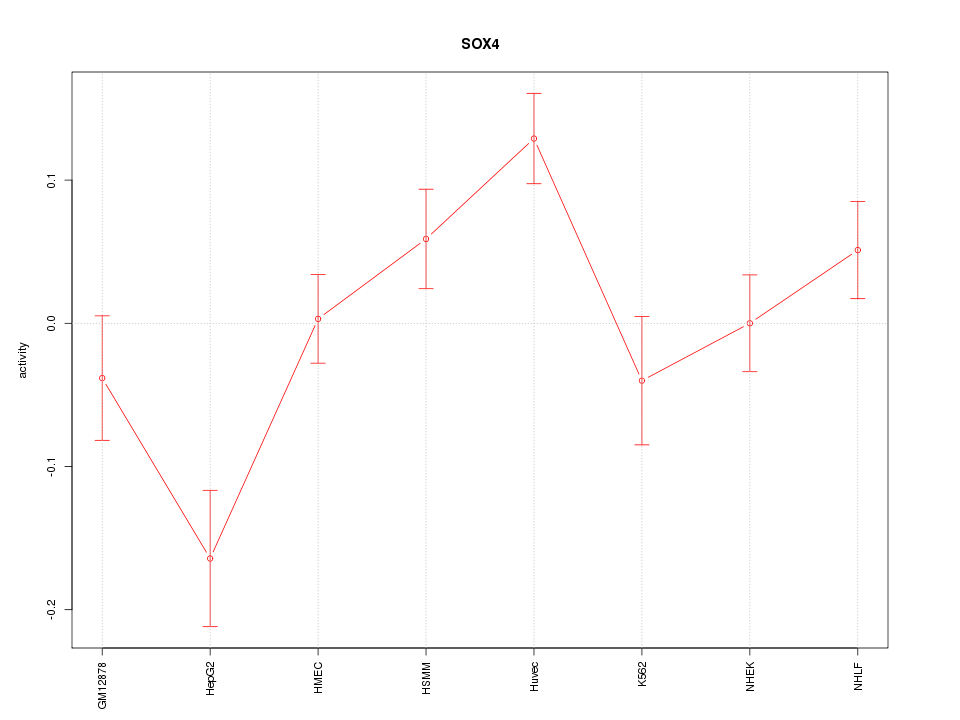
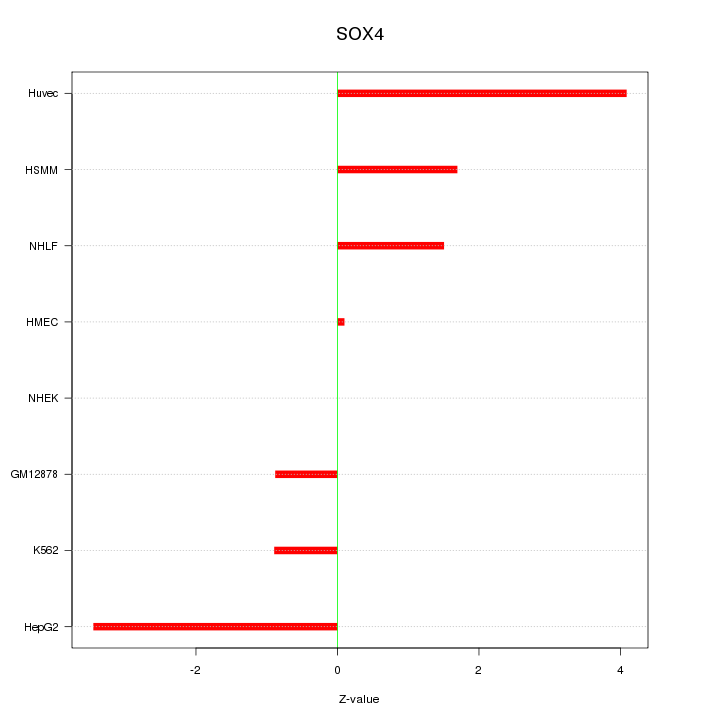
Motif ID: SOX4
Z-value: 2.102
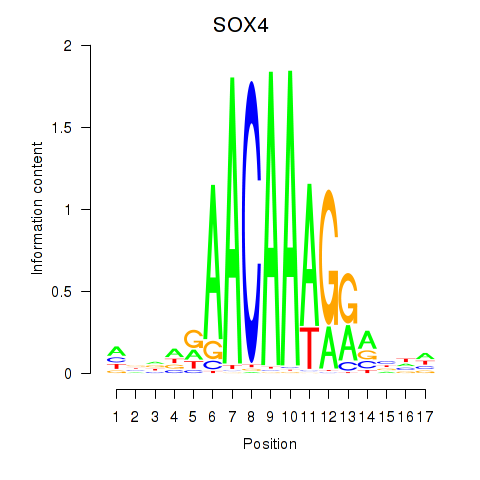
Transcription factors associated with SOX4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SOX4 | ENSG00000124766.4 | SOX4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 1.3 | 5.2 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 1.1 | 7.8 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.7 | 6.3 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.5 | 2.6 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.3 | 1.7 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 2.3 | GO:0003065 | regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.3 | 6.9 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.3 | 0.8 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.2 | 0.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 4.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.2 | 0.8 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.1 | 0.6 | GO:0034337 | RNA folding(GO:0034337) DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.4 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 2.4 | GO:1902749 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.1 | 1.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 2.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) minus-end-directed organelle transport along microtubule(GO:0072385) microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.9 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 1.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.4 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.1 | 0.8 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 6.1 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.1 | 3.1 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 3.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.8 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 7.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 2.2 | GO:0030323 | respiratory tube development(GO:0030323) |
| 0.0 | 0.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 6.7 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 2.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 2.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.1 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.7 | GO:0007049 | cell cycle(GO:0007049) |
| 0.0 | 3.2 | GO:0006260 | DNA replication(GO:0006260) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.4 | 1.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.3 | 1.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 0.5 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.2 | 2.3 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 1.9 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 13.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 3.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.3 | GO:0033655 | host(GO:0018995) host cell cytoplasm(GO:0030430) host cell part(GO:0033643) host intracellular part(GO:0033646) host cell cytoplasm part(GO:0033655) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 7.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 4.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 13.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 5.8 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.1 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.6 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 3.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 1.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 3.5 | GO:0015629 | actin cytoskeleton(GO:0015629) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.9 | 3.6 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.6 | 6.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 1.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 2.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 6.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 6.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.2 | 1.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 4.6 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 1.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.5 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 7.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.5 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 3.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 10.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 9.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |