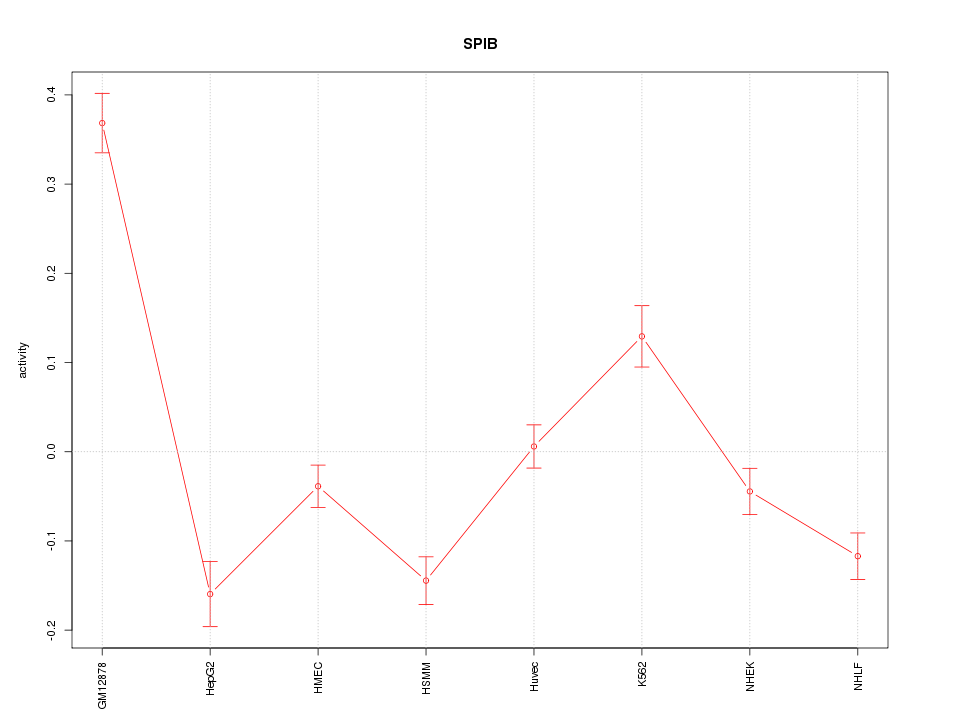
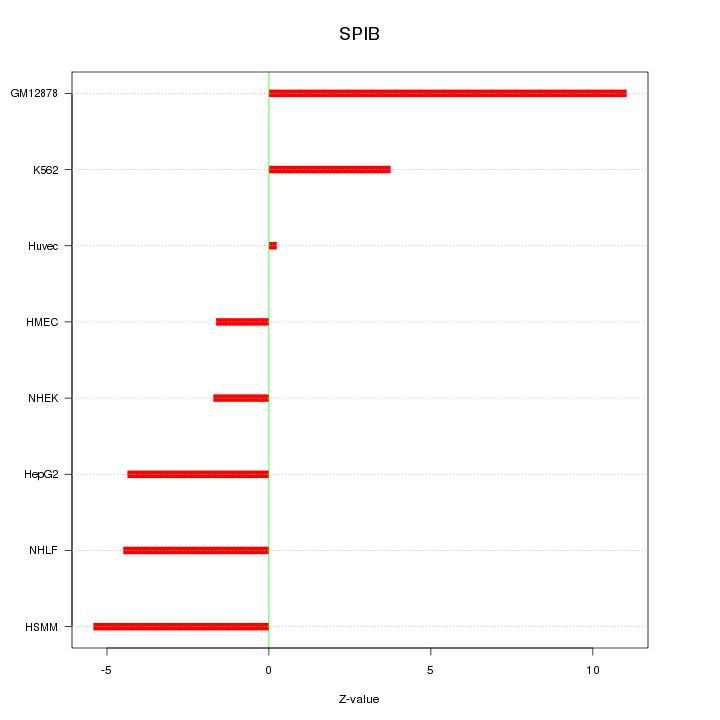
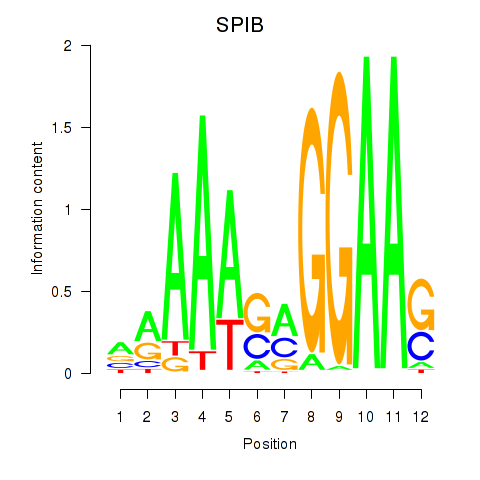
Motif ID: SPIB
Z-value: 5.129
Transcription factors associated with SPIB:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SPIB | ENSG00000269404.2 | SPIB |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.8 | GO:0042097 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 3.2 | 9.7 | GO:0043366 | beta selection(GO:0043366) |
| 3.0 | 8.9 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 2.0 | 8.2 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 1.9 | 7.8 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.8 | 7.4 | GO:0070527 | integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) platelet aggregation(GO:0070527) |
| 1.8 | 11.0 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 1.7 | 5.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 1.2 | 1.2 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 1.0 | 1.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) regulation of leukocyte degranulation(GO:0043300) negative regulation of leukocyte degranulation(GO:0043301) |
| 1.0 | 5.2 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 1.0 | 18.4 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.8 | 5.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.8 | 5.8 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.8 | 3.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.8 | 12.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.7 | 3.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.7 | 3.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.7 | 2.7 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.7 | 3.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.7 | 3.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.6 | 4.1 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.6 | 4.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.5 | 1.6 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.5 | 4.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.5 | 2.5 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.4 | 1.3 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.4 | 2.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.4 | 27.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.4 | 1.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) mitochondrial calcium ion homeostasis(GO:0051560) positive regulation of mitochondrial calcium ion concentration(GO:0051561) relaxation of skeletal muscle(GO:0090076) |
| 0.4 | 6.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.4 | 16.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.3 | 1.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 4.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.3 | 7.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 1.0 | GO:0007389 | pattern specification process(GO:0007389) |
| 0.3 | 2.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.3 | 4.3 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.3 | 5.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.3 | 1.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 8.8 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.3 | 0.9 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.3 | 3.5 | GO:0002639 | positive regulation of immunoglobulin production(GO:0002639) |
| 0.3 | 1.2 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.3 | 8.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.3 | 2.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.2 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.2 | 3.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.2 | 0.9 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.2 | 29.3 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.2 | 0.7 | GO:0019056 | modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 2.4 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.2 | 0.8 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.2 | 3.4 | GO:0051250 | negative regulation of lymphocyte activation(GO:0051250) |
| 0.2 | 3.0 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.2 | 7.5 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.2 | 15.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.2 | 1.1 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.2 | 1.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 2.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 6.9 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 0.6 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.7 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.1 | 0.5 | GO:0045349 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) negative regulation of toll-like receptor signaling pathway(GO:0034122) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 1.1 | GO:0010737 | protein kinase A signaling(GO:0010737) regulation of protein kinase A signaling(GO:0010738) protein maturation by protein folding(GO:0022417) |
| 0.1 | 1.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 1.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 3.4 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.1 | 1.3 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.1 | 1.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.3 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.1 | 1.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 0.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 1.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 4.6 | GO:0006914 | autophagy(GO:0006914) |
| 0.1 | 1.3 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 7.9 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.1 | 5.2 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 5.5 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 1.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 3.5 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.1 | 0.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.1 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) |
| 0.1 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 6.7 | GO:0050776 | regulation of immune response(GO:0050776) |
| 0.1 | 1.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 0.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 5.4 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 1.9 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.0 | 0.2 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.7 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 1.0 | GO:0044349 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.0 | 2.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.4 | GO:0043558 | regulation of translation in response to stress(GO:0043555) regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 1.0 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.7 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 3.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 2.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 2.0 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.0 | 0.5 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 6.8 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.8 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 3.0 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 5.1 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.4 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 7.2 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 8.0 | GO:0009968 | negative regulation of signal transduction(GO:0009968) |
| 0.0 | 0.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.4 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.5 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 0.2 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.2 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 1.6 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 3.1 | GO:0016049 | cell growth(GO:0016049) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.9 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.4 | GO:0006415 | translational termination(GO:0006415) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.4 | GO:0019815 | immunoglobulin complex(GO:0019814) B cell receptor complex(GO:0019815) |
| 1.9 | 5.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 1.3 | 8.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.8 | 16.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.8 | 4.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.6 | 5.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.6 | 2.4 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.6 | 6.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 7.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.5 | 3.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 2.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 3.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 2.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 14.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 6.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 0.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 1.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 7.3 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 1.4 | GO:0060076 | postsynaptic density(GO:0014069) excitatory synapse(GO:0060076) postsynaptic specialization(GO:0099572) |
| 0.1 | 5.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 4.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 7.9 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.1 | 1.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.2 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.1 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 32.5 | GO:0016023 | cytoplasmic, membrane-bounded vesicle(GO:0016023) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 3.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 7.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 3.4 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 4.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 127.4 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 4.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 3.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 3.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 14.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 6.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 9.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 5.5 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 1.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 32.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 2.1 | 6.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 1.9 | 7.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 1.7 | 20.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 1.2 | 3.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.1 | 12.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.9 | 4.5 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.8 | 7.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.7 | 2.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) |
| 0.7 | 7.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.7 | 2.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.7 | 3.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) |
| 0.6 | 3.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.6 | 1.9 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.6 | 2.4 | GO:0048039 | succinate dehydrogenase activity(GO:0000104) oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) ubiquinone binding(GO:0048039) |
| 0.6 | 5.1 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.5 | 12.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.5 | 8.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.5 | 2.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.5 | 6.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.5 | 5.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 2.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.4 | 9.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 5.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.4 | 7.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 7.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 11.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.3 | 4.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 1.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 2.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.3 | 1.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 3.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 2.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 3.2 | GO:0016896 | exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.2 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 7.8 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.2 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 3.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 2.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 5.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 4.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 8.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 3.9 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 4.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 5.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 5.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.6 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.1 | 1.1 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 1.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 2.7 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 0.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.9 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 11.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 1.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.1 | 5.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 23.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.6 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 4.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 4.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 16.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.3 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 4.0 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 3.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 5.7 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 1.3 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 1.0 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 2.5 | GO:0010843 | obsolete promoter binding(GO:0010843) |
| 0.0 | 3.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0009975 | cyclase activity(GO:0009975) phosphorus-oxygen lyase activity(GO:0016849) |
| 0.0 | 17.1 | GO:0004871 | signal transducer activity(GO:0004871) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.9 | GO:0019901 | protein kinase binding(GO:0019901) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 41.9 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 7.5 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.0 | 2.1 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.9 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 2.4 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |