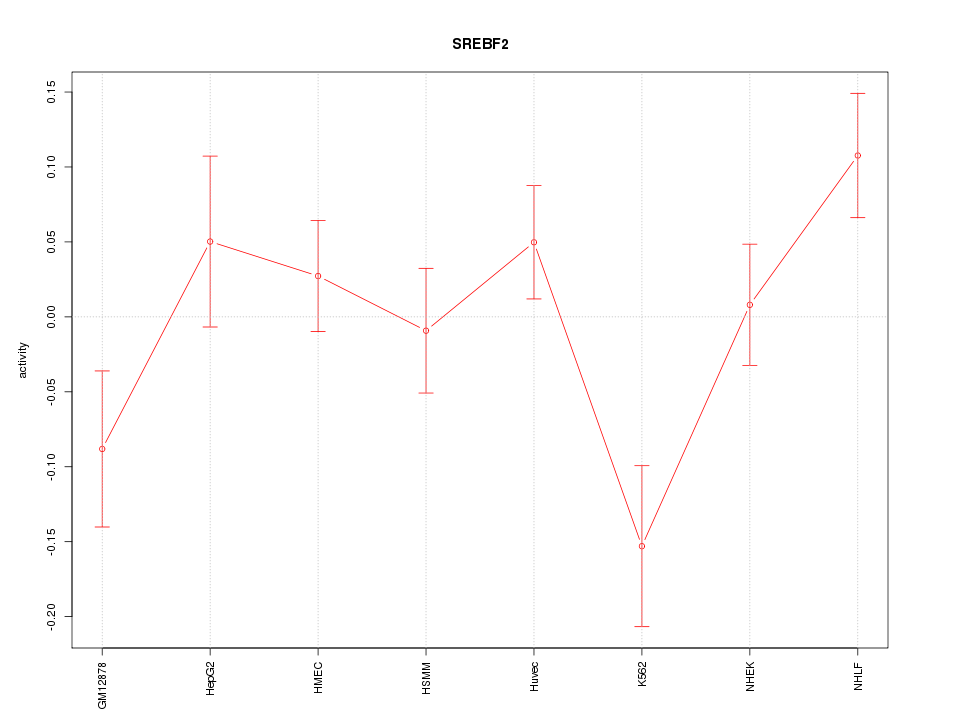
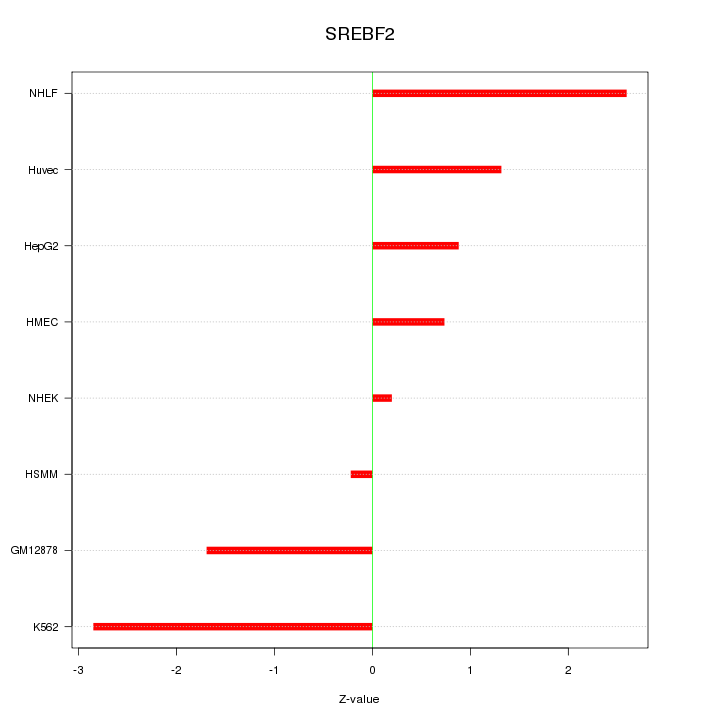
Motif ID: SREBF2
Z-value: 1.615
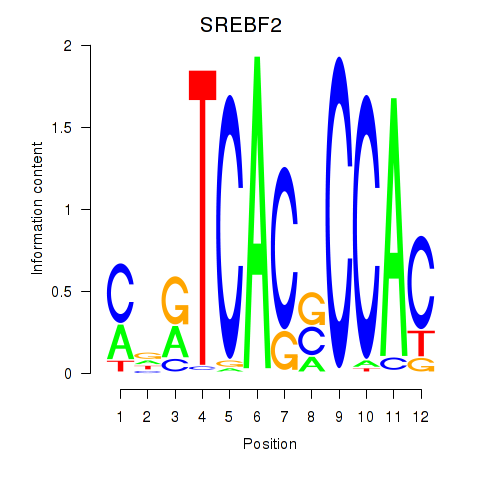
Transcription factors associated with SREBF2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SREBF2 | ENSG00000198911.7 | SREBF2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.7 | 2.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.6 | 2.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.6 | 4.8 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.6 | 5.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.5 | 1.9 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.4 | 3.0 | GO:0046618 | drug transmembrane transport(GO:0006855) tetracycline transport(GO:0015904) drug export(GO:0046618) |
| 0.3 | 5.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 1.1 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.1 | 2.8 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 1.6 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 0.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 2.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.3 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 | 3.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.7 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 2.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 2.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 4.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 5.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 7.7 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 5.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 2.6 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.7 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.7 | 5.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.6 | 3.0 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.6 | 5.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.5 | 4.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 7.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.7 | GO:0004741 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 3.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.2 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 2.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0051015 | actin filament binding(GO:0051015) |