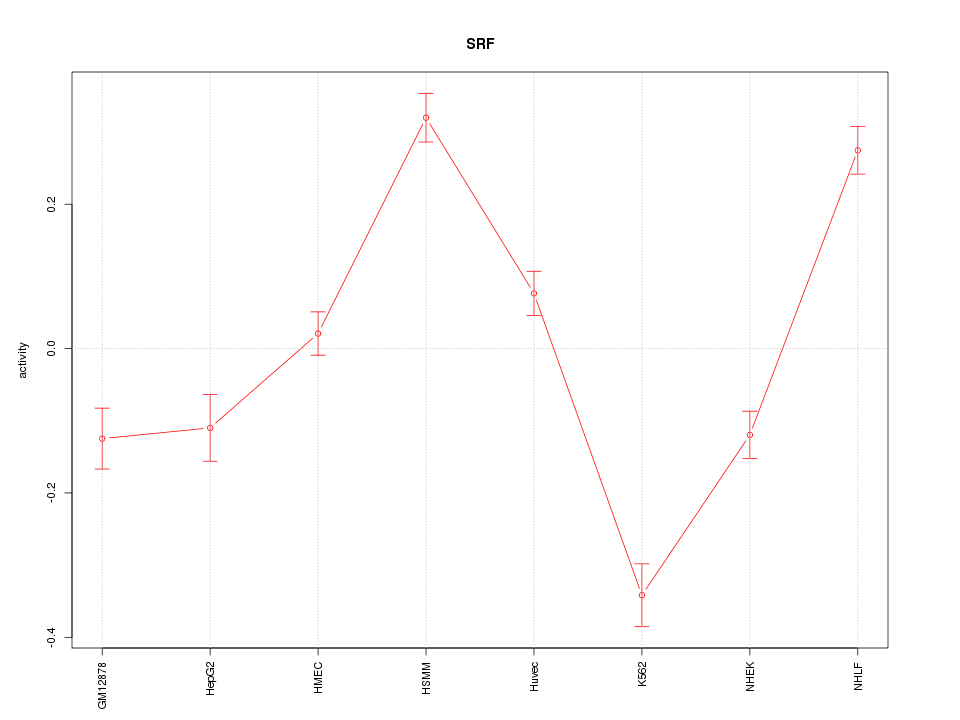
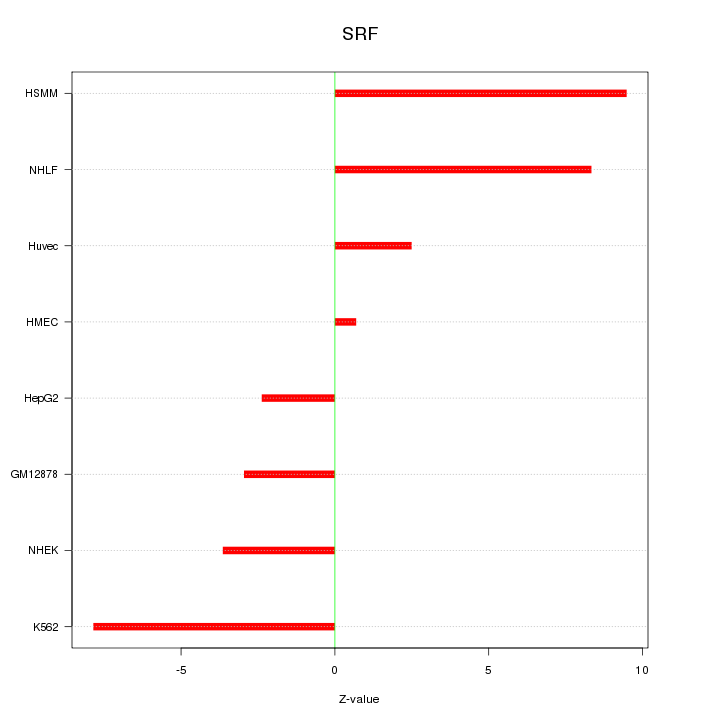
Motif ID: SRF
Z-value: 5.652
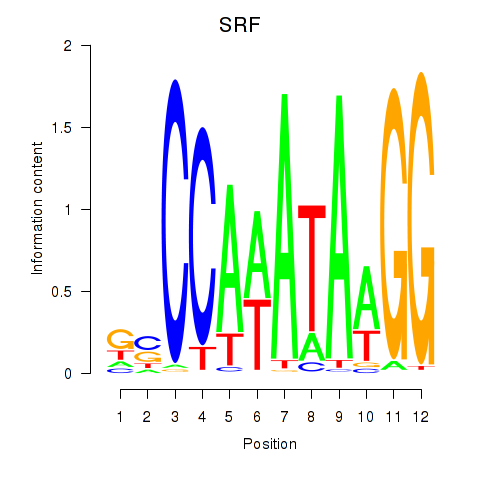
Transcription factors associated with SRF:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SRF | ENSG00000112658.6 | SRF |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 29.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 3.7 | 18.6 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.5 | 24.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 2.0 | 6.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 1.7 | 5.0 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 1.6 | 6.4 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.2 | 3.7 | GO:0060574 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 1.1 | 4.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.1 | 8.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 1.0 | 3.1 | GO:0014831 | intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.9 | 13.2 | GO:0000052 | citrulline metabolic process(GO:0000052) arginine catabolic process(GO:0006527) |
| 0.7 | 2.9 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.6 | 3.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.6 | 1.7 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.6 | 2.2 | GO:0060839 | radial pattern formation(GO:0009956) lymphatic endothelial cell fate commitment(GO:0060838) endothelial cell fate commitment(GO:0060839) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.5 | 5.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.5 | 1.6 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.5 | 2.0 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.5 | 6.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.4 | 6.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.4 | 3.8 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.4 | 35.1 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.4 | 5.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.4 | 11.6 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.3 | 12.3 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.3 | 26.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.3 | 6.4 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.3 | 2.3 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.3 | 2.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 1.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 2.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 | 3.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 4.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.2 | 4.2 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.2 | 1.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 43.5 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.1 | 6.9 | GO:0051318 | G1 phase(GO:0051318) |
| 0.1 | 19.3 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 4.5 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 4.3 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 11.1 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.3 | GO:0006740 | glyoxylate cycle(GO:0006097) NADPH regeneration(GO:0006740) |
| 0.1 | 5.1 | GO:0001764 | neuron migration(GO:0001764) |
| 0.1 | 9.3 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.1 | 1.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 8.8 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.1 | 1.5 | GO:0009067 | aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.1 | 1.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 2.2 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 9.3 | GO:0034329 | cell junction assembly(GO:0034329) |
| 0.0 | 0.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 4.3 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 1.2 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 24.7 | GO:0044255 | cellular lipid metabolic process(GO:0044255) |
| 0.0 | 7.0 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.8 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.6 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 27.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.3 | 23.4 | GO:0031430 | M band(GO:0031430) |
| 1.8 | 5.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.5 | 4.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.2 | 23.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.2 | 32.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.8 | 6.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.8 | 48.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.3 | 14.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 6.0 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.2 | 5.2 | GO:0036379 | myofilament(GO:0036379) |
| 0.2 | 2.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 25.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 1.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 6.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.9 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 7.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.9 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 2.1 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 6.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 19.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.0 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 32.7 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 1.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) protein acetyltransferase complex(GO:0031248) acetyltransferase complex(GO:1902493) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 7.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 22.6 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 3.1 | 24.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 2.6 | 13.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 1.2 | 5.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.9 | 4.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.7 | 22.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.6 | 8.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.6 | 4.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.6 | 53.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 7.3 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.5 | 1.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.5 | 3.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.5 | 2.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.5 | 6.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 2.0 | GO:0050833 | pyruvate secondary active transmembrane transporter activity(GO:0005477) pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 5.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.5 | 4.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.4 | 1.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 26.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.4 | 2.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 6.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 3.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 0.6 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.2 | 4.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 2.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 3.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 3.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 3.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 5.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 6.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 10.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 26.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.9 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 3.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 5.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 9.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 11.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 16.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 3.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 17.6 | GO:0005524 | ATP binding(GO:0005524) |
| 0.0 | 7.3 | GO:0016462 | pyrophosphatase activity(GO:0016462) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 44.7 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.0 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.2 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |