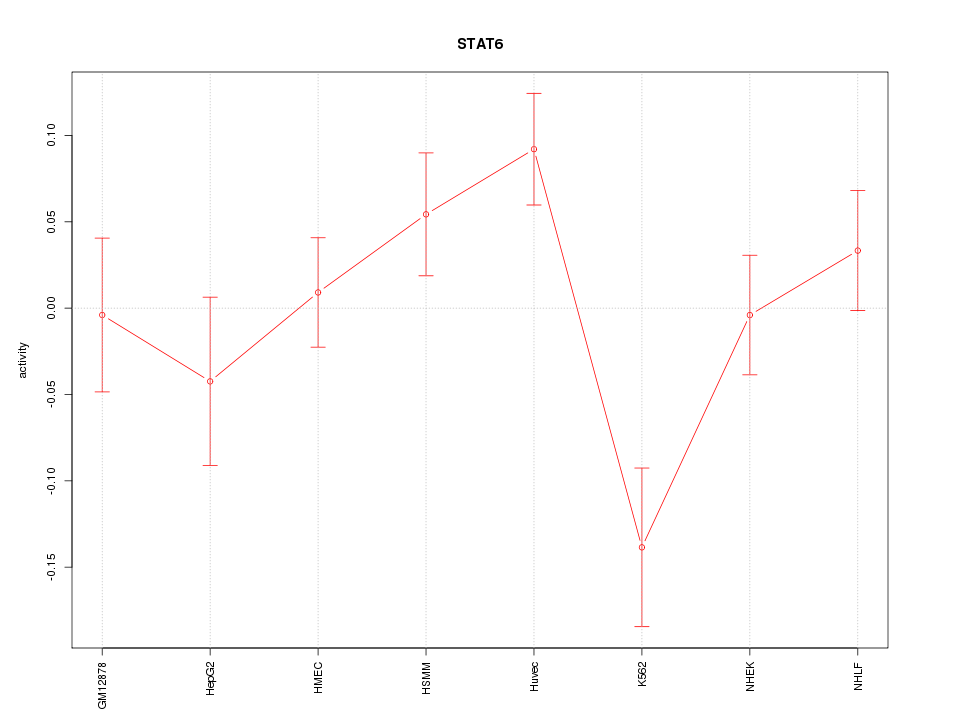
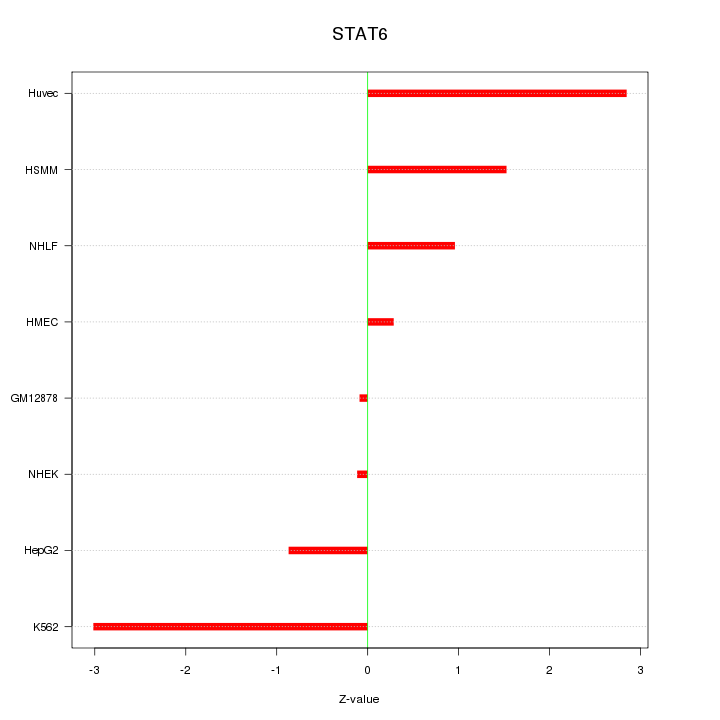
Motif ID: STAT6
Z-value: 1.632
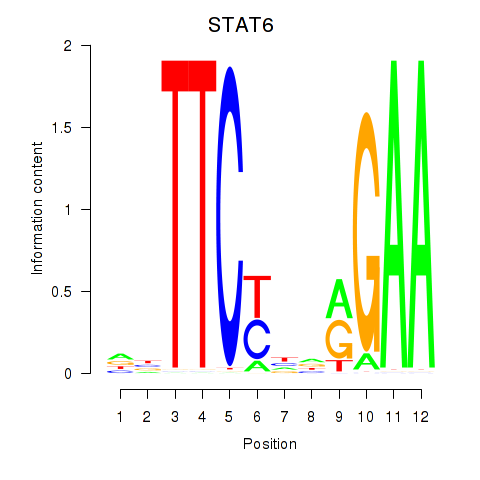
Transcription factors associated with STAT6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| STAT6 | ENSG00000166888.6 | STAT6 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.0 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.5 | 1.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.4 | 1.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 0.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.6 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.2 | 1.7 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 | 1.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 2.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.7 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.3 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 1.7 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.1 | 3.2 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.4 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 1.3 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.2 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.7 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 1.0 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 0.7 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 3.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.4 | GO:0009584 | detection of visible light(GO:0009584) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 5.0 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.4 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.6 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.8 | GO:0030335 | positive regulation of cell migration(GO:0030335) positive regulation of cell motility(GO:2000147) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 3.0 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.1 | 3.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 3.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 4.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 2.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.5 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 2.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 5.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.4 | 1.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 1.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 1.0 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.3 | 1.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 4.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 3.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 1.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.6 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 3.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.2 | GO:0046980 | oligopeptide-transporting ATPase activity(GO:0015421) tapasin binding(GO:0046980) |
| 0.0 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |