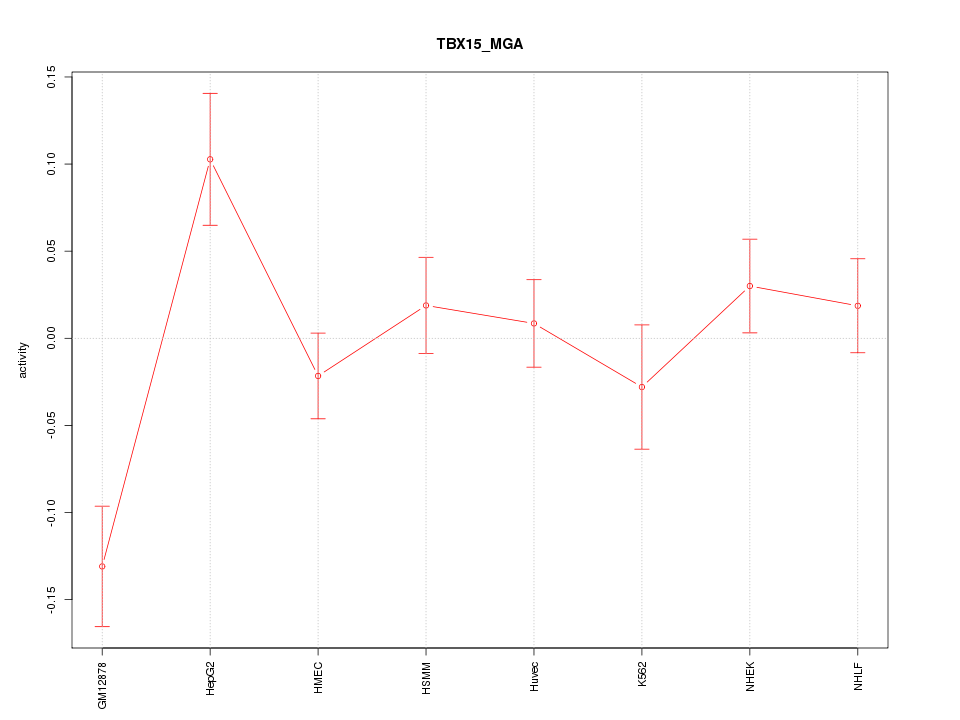
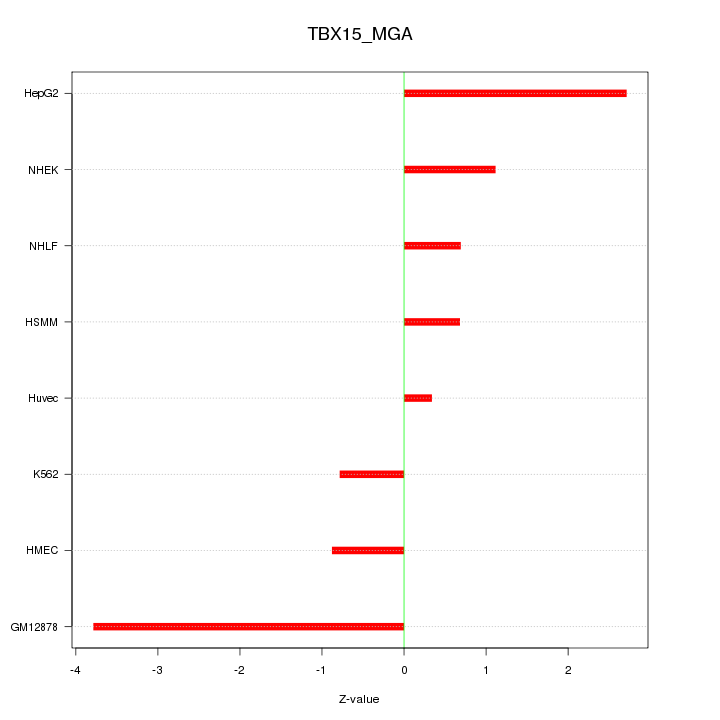
Motif ID: TBX15_MGA
Z-value: 1.782
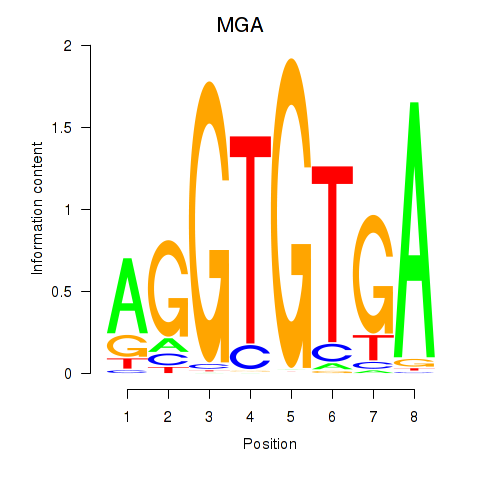
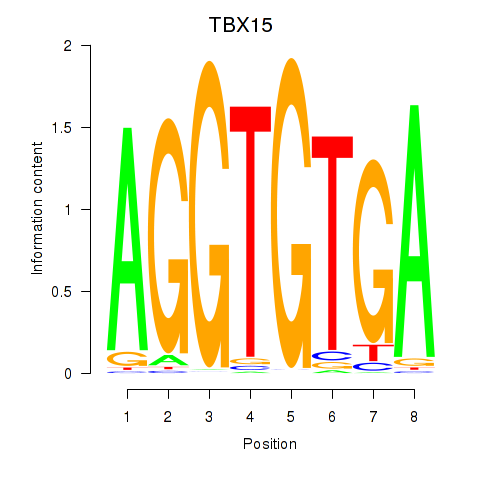
Transcription factors associated with TBX15_MGA:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| MGA | ENSG00000174197.12 | MGA |
| TBX15 | ENSG00000092607.9 | TBX15 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0001705 | ectoderm formation(GO:0001705) central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.8 | 3.3 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.7 | 2.1 | GO:0003176 | aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) notochord formation(GO:0014028) |
| 0.6 | 1.8 | GO:2000910 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.5 | 4.8 | GO:0030277 | epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.5 | 2.0 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.5 | 0.5 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.4 | 1.7 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.4 | 3.9 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.4 | 3.4 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.4 | 1.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.3 | 1.4 | GO:0034442 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.3 | 2.0 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 | 1.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.3 | 2.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 | 1.8 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.3 | 1.5 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.3 | 1.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 1.2 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.2 | 0.7 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.2 | 1.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.8 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 1.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 3.0 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.1 | 0.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 5.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.4 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) muscle hyperplasia(GO:0014900) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.1 | 0.7 | GO:0003070 | age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.1 | 0.4 | GO:0002575 | basophil chemotaxis(GO:0002575) |
| 0.1 | 1.7 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.4 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 0.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 3.7 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.1 | GO:0070849 | response to epidermal growth factor(GO:0070849) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 1.9 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.6 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.6 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.6 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 2.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.3 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.1 | 2.4 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.9 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.3 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 | 0.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.2 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.1 | 0.2 | GO:0055005 | pulmonary myocardium development(GO:0003350) ventricular cardiac myofibril assembly(GO:0055005) left/right pattern formation(GO:0060972) |
| 0.1 | 1.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.5 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.0 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 1.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 1.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.4 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.9 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.9 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 1.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.1 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 1.8 | GO:0071772 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.1 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 2.4 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 3.0 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.1 | GO:0061245 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.3 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.2 | GO:0045727 | positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.3 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 1.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 5.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 5.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.0 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 3.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 2.6 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 3.6 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 1.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.3 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 2.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.6 | 2.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.5 | 1.5 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.5 | 2.5 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.5 | 3.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.4 | 1.8 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 1.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.4 | 1.8 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.4 | 1.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 3.4 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.3 | 1.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 1.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 1.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 0.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 3.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 3.6 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.2 | 0.7 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.2 | 2.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 2.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 3.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 2.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.9 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 0.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.7 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 1.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 7.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.2 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 5.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 3.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0004461 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 3.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.2 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.0 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0000049 | tRNA binding(GO:0000049) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |