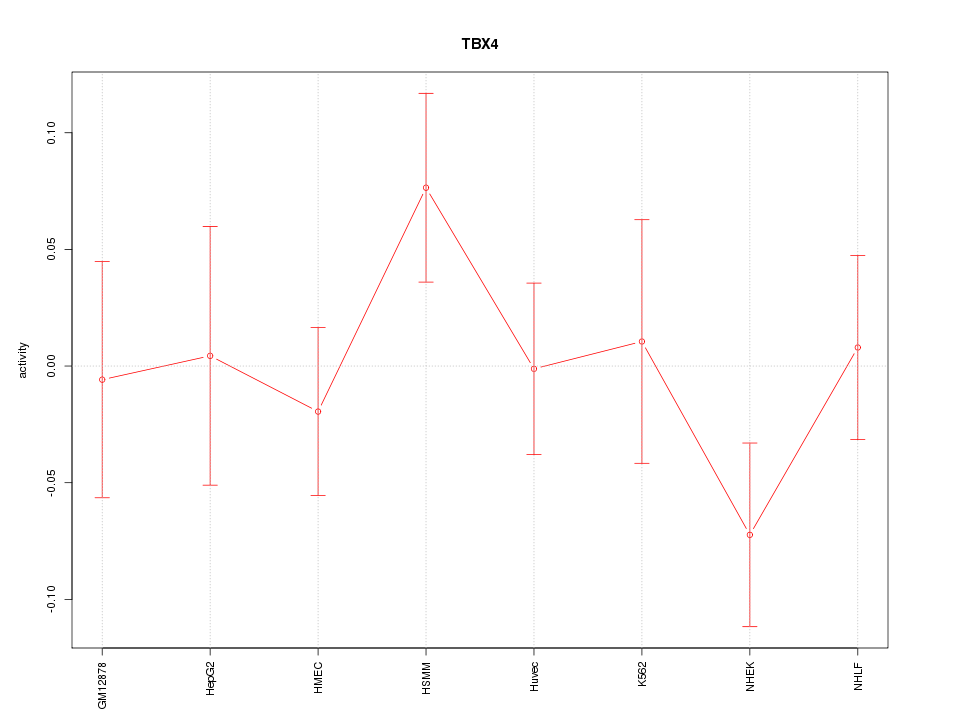
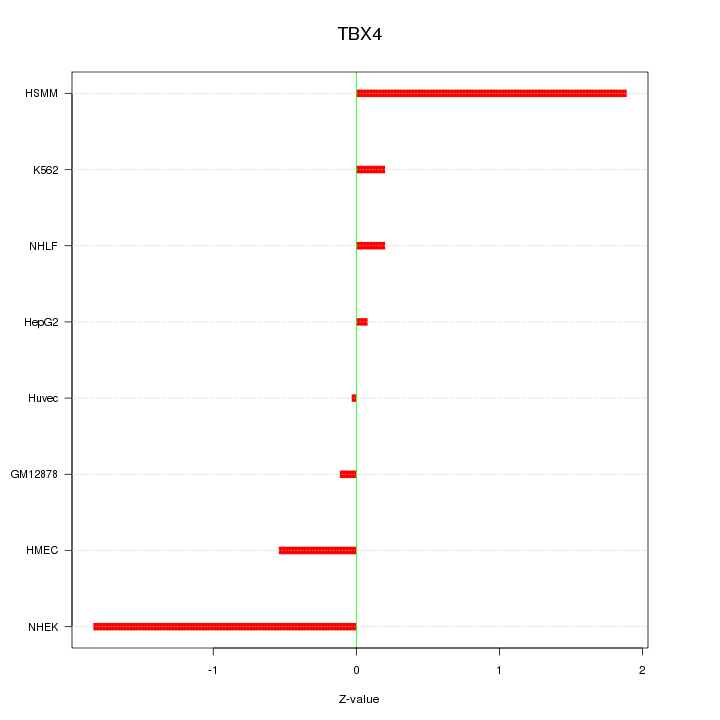
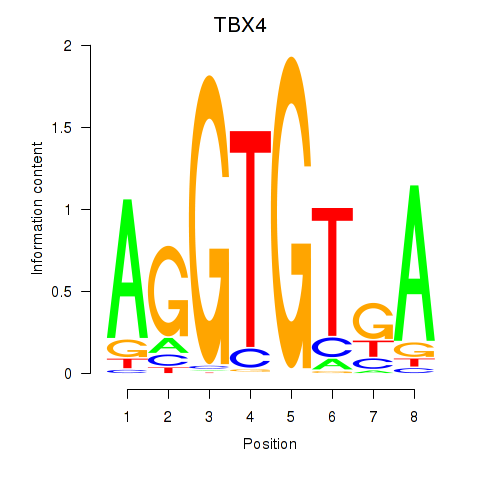
Motif ID: TBX4
Z-value: 0.958
Transcription factors associated with TBX4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| TBX4 | ENSG00000121075.5 | TBX4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 0.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.8 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 1.0 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.5 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.6 | GO:0048268 | negative regulation of receptor-mediated endocytosis(GO:0048261) clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:1902170 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.1 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0010535 | regulation of activation of JAK2 kinase activity(GO:0010534) positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 | 4.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 3.0 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 0.3 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 1.2 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 4.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 4.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |