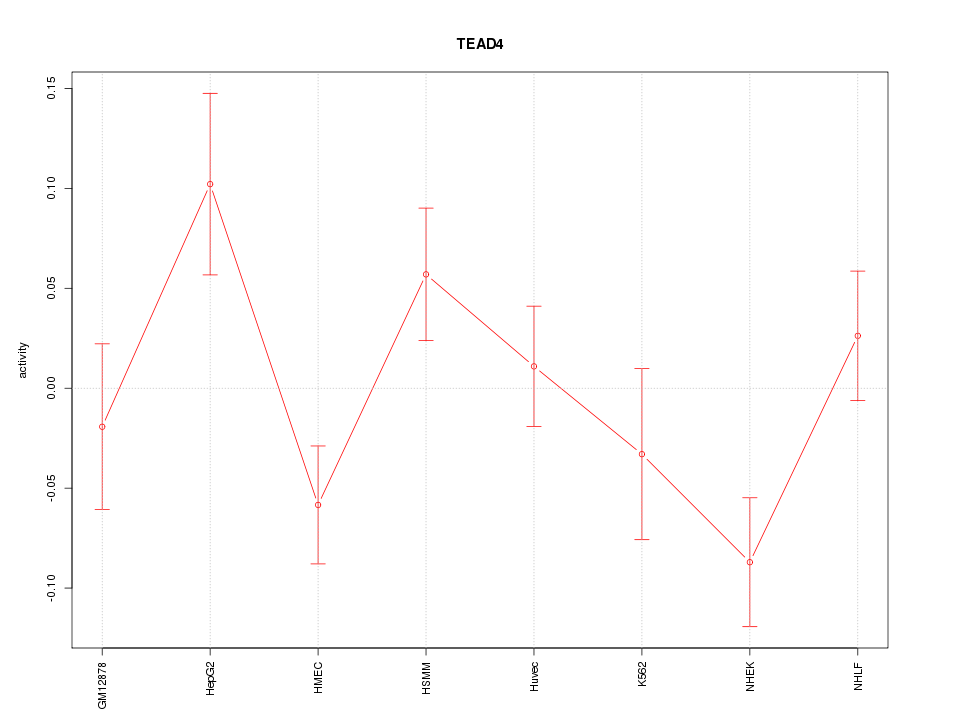
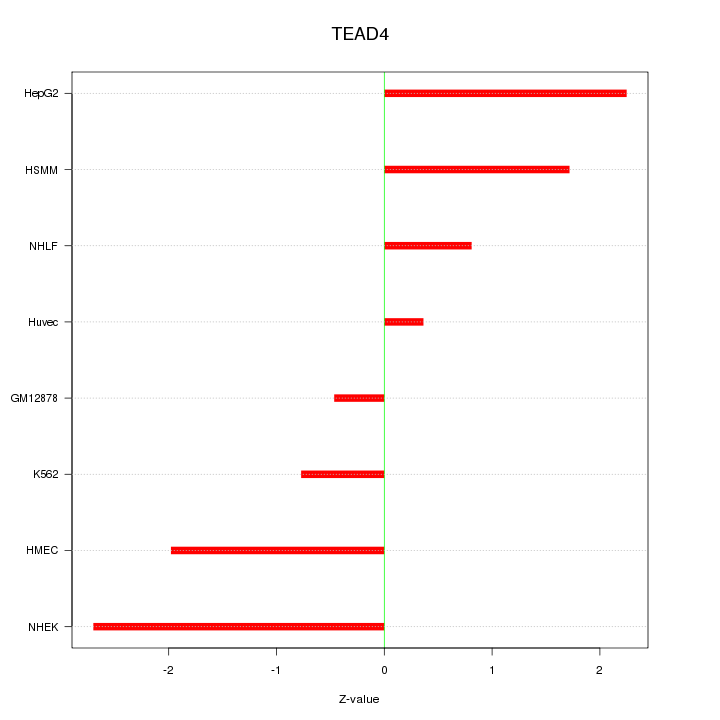
Motif ID: TEAD4
Z-value: 1.613
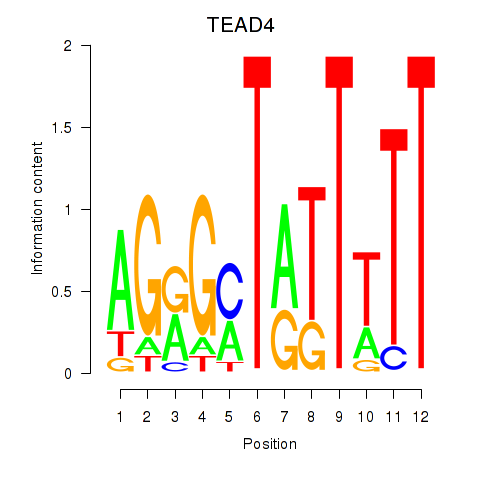
Transcription factors associated with TEAD4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| TEAD4 | ENSG00000197905.4 | TEAD4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.5 | 1.5 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.4 | 4.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.3 | 0.8 | GO:0031034 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 2.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 11.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.5 | GO:0014831 | intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.1 | 1.4 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.5 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 | 1.3 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 0.3 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.7 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 1.5 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.1 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.0 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.2 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 1.4 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.1 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.0 | 0.7 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.3 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 3.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.9 | GO:0051923 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) sulfation(GO:0051923) |
| 0.0 | 3.8 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.1 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 3.3 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.0 | GO:1900181 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein localization to nucleus(GO:1900181) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 1.0 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 1.5 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.1 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.8 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 1.0 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) minus-end-directed organelle transport along microtubule(GO:0072385) microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 1.2 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.2 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.0 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.2 | 2.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 2.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 26.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.0 | GO:0033655 | host(GO:0018995) host cell cytoplasm(GO:0030430) host cell part(GO:0033643) host intracellular part(GO:0033646) host cell cytoplasm part(GO:0033655) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0047042 | androsterone dehydrogenase (B-specific) activity(GO:0047042) indanol dehydrogenase activity(GO:0047718) |
| 0.2 | 11.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 1.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 4.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 1.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.3 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) alpha-catenin binding(GO:0045294) |
| 0.1 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 3.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.1 | 1.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 3.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 4.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 1.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.7 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 2.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 3.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 1.1 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0015254 | urea transmembrane transporter activity(GO:0015204) glycerol channel activity(GO:0015254) |
| 0.0 | 1.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |