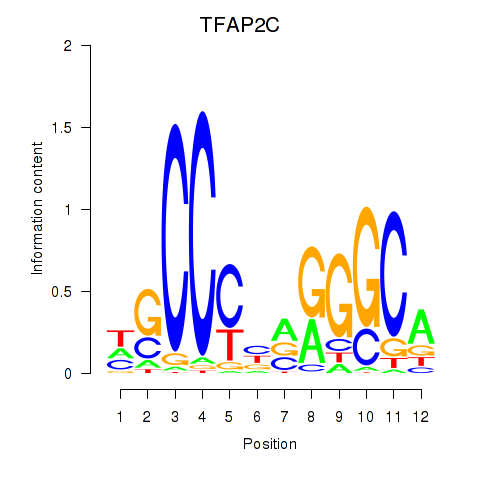
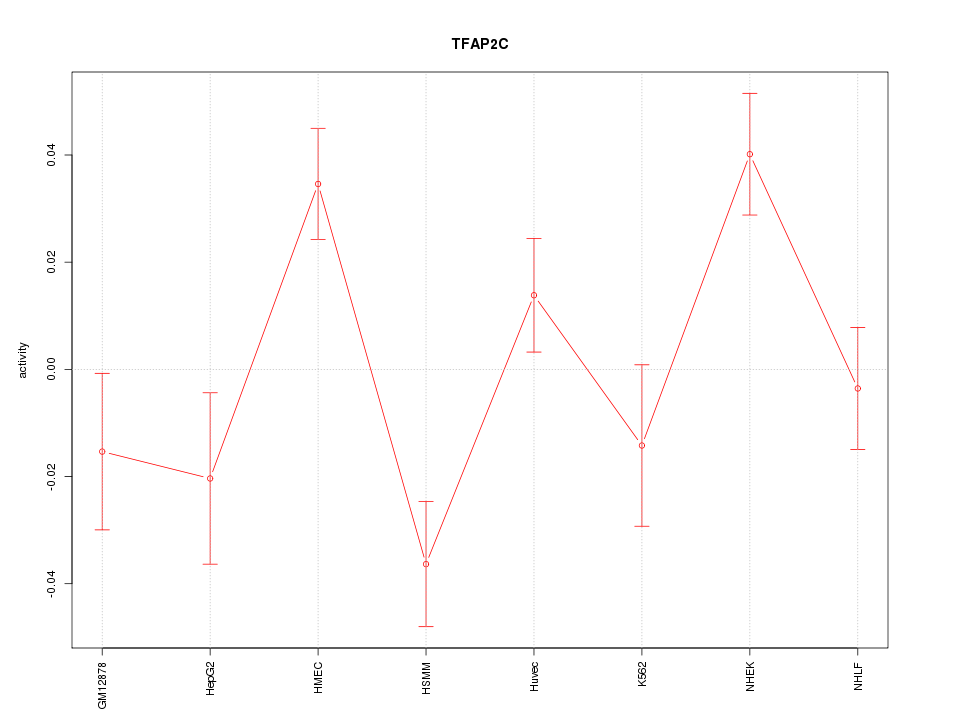
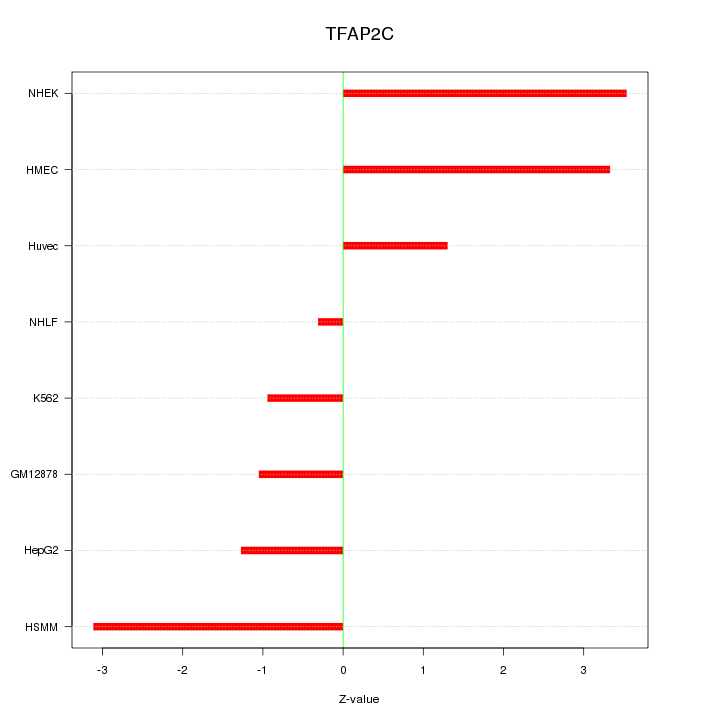
| 1.0 |
3.9 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.8 |
2.3 |
GO:0003308 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.7 |
3.0 |
GO:0060649 |
nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.7 |
2.2 |
GO:0019747 |
regulation of isoprenoid metabolic process(GO:0019747) |
| 0.7 |
2.2 |
GO:0003171 |
atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.7 |
2.0 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.6 |
1.8 |
GO:0046521 |
sphingoid catabolic process(GO:0046521) |
| 0.6 |
1.8 |
GO:0009648 |
photoperiodism(GO:0009648) |
| 0.5 |
1.8 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 |
0.9 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.4 |
1.3 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.4 |
1.3 |
GO:0010701 |
positive regulation of chronic inflammatory response(GO:0002678) positive regulation of norepinephrine secretion(GO:0010701) |
| 0.4 |
1.3 |
GO:0010837 |
regulation of keratinocyte proliferation(GO:0010837) |
| 0.4 |
2.1 |
GO:0042574 |
retinal metabolic process(GO:0042574) |
| 0.4 |
2.4 |
GO:0060263 |
regulation of respiratory burst(GO:0060263) |
| 0.4 |
1.2 |
GO:0061364 |
response to cortisol(GO:0051414) apoptotic process involved in luteolysis(GO:0061364) |
| 0.4 |
0.4 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.4 |
0.8 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.4 |
1.9 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.4 |
1.1 |
GO:0034205 |
beta-amyloid formation(GO:0034205) |
| 0.4 |
0.4 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.3 |
1.0 |
GO:0002541 |
activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.3 |
2.4 |
GO:0010591 |
regulation of lamellipodium assembly(GO:0010591) |
| 0.3 |
5.1 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.3 |
1.0 |
GO:0048627 |
myoblast development(GO:0048627) |
| 0.3 |
1.3 |
GO:0042335 |
cuticle development(GO:0042335) |
| 0.3 |
1.0 |
GO:0046495 |
nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.3 |
1.6 |
GO:0033484 |
nitric oxide homeostasis(GO:0033484) |
| 0.3 |
1.5 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.3 |
1.1 |
GO:0060406 |
positive regulation of penile erection(GO:0060406) |
| 0.3 |
0.8 |
GO:0046368 |
GDP-L-fucose biosynthetic process(GO:0042350) 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 |
1.0 |
GO:0032849 |
positive regulation of cellular pH reduction(GO:0032849) |
| 0.2 |
1.0 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.2 |
7.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.2 |
1.1 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.2 |
0.7 |
GO:0071476 |
response to vitamin B1(GO:0010266) cellular hypotonic response(GO:0071476) |
| 0.2 |
1.3 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 |
0.2 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.2 |
1.1 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 |
0.6 |
GO:0021769 |
orbitofrontal cortex development(GO:0021769) ventricular zone neuroblast division(GO:0021847) fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.2 |
1.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.2 |
0.6 |
GO:0050917 |
sensory perception of umami taste(GO:0050917) |
| 0.2 |
0.6 |
GO:0060054 |
positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 |
1.2 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 |
0.6 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 |
0.8 |
GO:0022614 |
membrane to membrane docking(GO:0022614) |
| 0.2 |
1.0 |
GO:0032933 |
SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 |
1.1 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 |
0.4 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 |
0.3 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 |
0.7 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 |
0.5 |
GO:0010248 |
establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.2 |
0.7 |
GO:0009698 |
phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.2 |
0.5 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.2 |
0.2 |
GO:0048635 |
negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle organ development(GO:0048635) negative regulation of muscle tissue development(GO:1901862) |
| 0.2 |
0.5 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 |
1.3 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.2 |
0.6 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 |
4.6 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 |
2.4 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.2 |
0.6 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.1 |
0.7 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 |
0.4 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 |
0.6 |
GO:0045338 |
farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 |
0.4 |
GO:0046984 |
regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.1 |
2.7 |
GO:0043616 |
keratinocyte proliferation(GO:0043616) |
| 0.1 |
2.7 |
GO:1901224 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 |
0.4 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.1 |
1.0 |
GO:2000109 |
macrophage apoptotic process(GO:0071888) regulation of macrophage apoptotic process(GO:2000109) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 |
1.8 |
GO:1901797 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.1 |
0.4 |
GO:0035283 |
central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 |
3.3 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 |
1.4 |
GO:0043497 |
regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 |
0.4 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.1 |
0.8 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 |
0.4 |
GO:0048549 |
positive regulation of pinocytosis(GO:0048549) |
| 0.1 |
0.4 |
GO:0032509 |
endosome transport via multivesicular body sorting pathway(GO:0032509) endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 |
0.9 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.1 |
0.2 |
GO:0021836 |
chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.1 |
3.0 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.1 |
1.1 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.1 |
0.4 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
1.0 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
0.2 |
GO:0032707 |
negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 |
1.2 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.1 |
0.6 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
0.5 |
GO:0019046 |
release from viral latency(GO:0019046) |
| 0.1 |
0.5 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 |
1.6 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.1 |
0.3 |
GO:0009153 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.1 |
0.3 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 |
0.1 |
GO:0043949 |
regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 |
0.3 |
GO:0032226 |
positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.1 |
0.2 |
GO:0090598 |
male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 |
1.1 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 |
0.2 |
GO:0032621 |
interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.1 |
0.2 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 |
0.4 |
GO:0072048 |
pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.1 |
0.4 |
GO:0045091 |
regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 |
0.6 |
GO:0006880 |
intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 |
0.3 |
GO:0006427 |
histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 |
0.4 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 |
0.7 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 |
0.9 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
0.5 |
GO:0006591 |
ornithine metabolic process(GO:0006591) |
| 0.1 |
0.2 |
GO:0040014 |
regulation of multicellular organism growth(GO:0040014) |
| 0.1 |
0.1 |
GO:0060585 |
regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 |
0.5 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.1 |
0.4 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 |
0.9 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 |
0.3 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.1 |
0.8 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 |
0.5 |
GO:0001960 |
negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.1 |
0.6 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 |
1.4 |
GO:1902668 |
negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 |
0.3 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 |
1.8 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.1 |
0.4 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 |
0.3 |
GO:0006015 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 |
0.3 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.3 |
GO:0021873 |
forebrain neuroblast division(GO:0021873) neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.1 |
0.3 |
GO:0005997 |
xylulose metabolic process(GO:0005997) |
| 0.1 |
0.1 |
GO:0034086 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 |
0.6 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.1 |
0.5 |
GO:0042159 |
lipoprotein catabolic process(GO:0042159) |
| 0.1 |
10.6 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 |
0.2 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.1 |
0.6 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.1 |
0.6 |
GO:1901998 |
tetracycline transport(GO:0015904) antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.1 |
0.8 |
GO:0055015 |
ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 |
0.2 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.1 |
1.3 |
GO:0051155 |
positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.1 |
0.1 |
GO:0033088 |
immature T cell proliferation in thymus(GO:0033080) regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 |
0.2 |
GO:0006788 |
heme oxidation(GO:0006788) |
| 0.1 |
0.4 |
GO:0046952 |
ketone body catabolic process(GO:0046952) |
| 0.1 |
2.7 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.1 |
0.3 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 |
0.1 |
GO:0031573 |
intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 |
0.2 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 |
1.2 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 |
0.1 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 |
0.6 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 |
0.4 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.1 |
0.3 |
GO:0018352 |
protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 |
0.1 |
GO:0031558 |
obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.1 |
0.2 |
GO:0042759 |
fatty acid elongation, saturated fatty acid(GO:0019367) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.2 |
GO:0051547 |
regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 |
0.7 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 |
0.5 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.1 |
GO:0010565 |
regulation of cellular ketone metabolic process(GO:0010565) |
| 0.1 |
0.2 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 |
0.3 |
GO:0034204 |
lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 |
0.5 |
GO:0030041 |
actin filament polymerization(GO:0030041) |
| 0.1 |
1.8 |
GO:0060444 |
branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.1 |
0.1 |
GO:0008356 |
asymmetric cell division(GO:0008356) |
| 0.1 |
0.3 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
0.9 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.1 |
0.2 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 |
0.5 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 |
0.1 |
GO:0060526 |
prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.1 |
0.1 |
GO:1902667 |
regulation of axon extension involved in axon guidance(GO:0048841) regulation of axon guidance(GO:1902667) |
| 0.1 |
0.1 |
GO:1903170 |
negative regulation of ion transmembrane transporter activity(GO:0032413) negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of calcium ion transmembrane transport(GO:1903170) negative regulation of cation channel activity(GO:2001258) |
| 0.1 |
0.2 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
1.1 |
GO:0035338 |
long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 |
0.2 |
GO:0001973 |
adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 |
0.2 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
0.3 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.1 |
0.5 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 |
0.3 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.1 |
0.3 |
GO:0050932 |
regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.1 |
0.1 |
GO:0042524 |
negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.1 |
0.2 |
GO:0042369 |
vitamin D catabolic process(GO:0042369) |
| 0.1 |
0.2 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
0.2 |
GO:0003148 |
outflow tract septum morphogenesis(GO:0003148) |
| 0.1 |
0.3 |
GO:0048241 |
epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) |
| 0.1 |
0.2 |
GO:0042268 |
regulation of cytolysis(GO:0042268) |
| 0.1 |
0.2 |
GO:0007143 |
female meiotic division(GO:0007143) |
| 0.1 |
0.2 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.1 |
0.3 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 |
0.2 |
GO:0006787 |
porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 |
0.6 |
GO:0010745 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 |
0.9 |
GO:0008088 |
axo-dendritic transport(GO:0008088) |
| 0.1 |
0.1 |
GO:2001021 |
negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.1 |
0.1 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 0.1 |
1.6 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 |
0.2 |
GO:0006542 |
glutamine biosynthetic process(GO:0006542) |
| 0.0 |
0.1 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
1.0 |
GO:0050927 |
positive regulation of positive chemotaxis(GO:0050927) |
| 0.0 |
0.1 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.0 |
1.1 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.0 |
1.1 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.1 |
GO:0046604 |
regulation of mitotic centrosome separation(GO:0046602) positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 |
0.1 |
GO:0003099 |
positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 |
0.1 |
GO:0061117 |
negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 |
0.2 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 |
0.7 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.5 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
1.0 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.6 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.1 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.3 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.3 |
GO:0045040 |
outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 |
0.1 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 |
1.0 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.1 |
GO:0016572 |
histone phosphorylation(GO:0016572) |
| 0.0 |
0.4 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 |
0.1 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.2 |
GO:0048532 |
anatomical structure arrangement(GO:0048532) |
| 0.0 |
0.4 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.4 |
GO:0046051 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 |
0.0 |
GO:0060180 |
regulation of female receptivity(GO:0045924) female mating behavior(GO:0060180) |
| 0.0 |
0.2 |
GO:0071025 |
RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 |
0.1 |
GO:0048262 |
determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.0 |
0.2 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 |
0.1 |
GO:0019853 |
L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 |
0.2 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 |
0.2 |
GO:0032495 |
response to muramyl dipeptide(GO:0032495) |
| 0.0 |
0.4 |
GO:0045652 |
regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 |
0.2 |
GO:0051877 |
pigment granule aggregation in cell center(GO:0051877) |
| 0.0 |
1.1 |
GO:0006626 |
protein targeting to mitochondrion(GO:0006626) |
| 0.0 |
0.1 |
GO:0032025 |
response to cobalt ion(GO:0032025) |
| 0.0 |
0.4 |
GO:0006743 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.1 |
GO:0002384 |
hepatic immune response(GO:0002384) |
| 0.0 |
0.1 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 |
0.8 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
0.4 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.5 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.2 |
GO:0060317 |
cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 |
6.3 |
GO:0008544 |
epidermis development(GO:0008544) |
| 0.0 |
0.1 |
GO:0071109 |
thalamus development(GO:0021794) superior temporal gyrus development(GO:0071109) |
| 0.0 |
0.2 |
GO:0002312 |
somatic recombination of immunoglobulin genes involved in immune response(GO:0002204) somatic diversification of immunoglobulins involved in immune response(GO:0002208) B cell activation involved in immune response(GO:0002312) immunoglobulin production involved in immunoglobulin mediated immune response(GO:0002381) isotype switching(GO:0045190) |
| 0.0 |
0.2 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
0.1 |
GO:0001807 |
type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 |
0.1 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.1 |
GO:0046668 |
retinal cell programmed cell death(GO:0046666) regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 |
0.5 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.1 |
GO:0043012 |
regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 |
0.4 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
1.0 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.2 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.1 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 |
0.3 |
GO:0042517 |
positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 |
0.2 |
GO:2001259 |
positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.0 |
0.1 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.0 |
0.1 |
GO:0007199 |
G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 |
0.4 |
GO:0010812 |
negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 |
2.4 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 |
2.3 |
GO:0051271 |
negative regulation of cellular component movement(GO:0051271) |
| 0.0 |
0.3 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.6 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
0.8 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.0 |
0.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.2 |
GO:0034287 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 |
0.8 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.2 |
GO:0048488 |
synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.3 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.0 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 |
0.2 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.1 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
1.9 |
GO:0007623 |
circadian rhythm(GO:0007623) |
| 0.0 |
0.1 |
GO:0070528 |
protein kinase C signaling(GO:0070528) |
| 0.0 |
0.1 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.0 |
0.2 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.3 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 |
0.2 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.2 |
GO:0043114 |
regulation of vascular permeability(GO:0043114) |
| 0.0 |
0.3 |
GO:0006984 |
ER-nucleus signaling pathway(GO:0006984) |
| 0.0 |
0.1 |
GO:0032929 |
negative regulation of interleukin-12 production(GO:0032695) negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.1 |
GO:0030517 |
negative regulation of axon extension(GO:0030517) |
| 0.0 |
0.4 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.1 |
GO:0031958 |
corticosteroid receptor signaling pathway(GO:0031958) glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 |
0.3 |
GO:0046718 |
viral entry into host cell(GO:0046718) |
| 0.0 |
0.2 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
2.1 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.0 |
1.4 |
GO:0042035 |
regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 |
0.2 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.0 |
0.2 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.1 |
GO:0034243 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 |
0.1 |
GO:0045885 |
obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 |
0.1 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 |
0.1 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.1 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 |
0.2 |
GO:0045010 |
actin nucleation(GO:0045010) |
| 0.0 |
0.2 |
GO:0010894 |
negative regulation of steroid biosynthetic process(GO:0010894) |
| 0.0 |
0.1 |
GO:0030260 |
entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 |
0.2 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.1 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.0 |
0.2 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.1 |
GO:0030858 |
positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.0 |
0.2 |
GO:0006885 |
regulation of pH(GO:0006885) |
| 0.0 |
0.6 |
GO:2000181 |
negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 |
0.1 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) cellular response to topologically incorrect protein(GO:0035967) |
| 0.0 |
0.3 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.1 |
GO:0071344 |
diphosphate metabolic process(GO:0071344) |
| 0.0 |
1.1 |
GO:0008064 |
regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.0 |
0.1 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 |
3.4 |
GO:0043123 |
positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 |
0.0 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
1.4 |
GO:0006921 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 |
0.5 |
GO:0036503 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 |
0.1 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.3 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.2 |
GO:1901070 |
GMP biosynthetic process(GO:0006177) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.0 |
0.1 |
GO:0034126 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) positive regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034126) |
| 0.0 |
0.1 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.2 |
GO:0006356 |
regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 |
0.0 |
GO:0030157 |
carbon dioxide transport(GO:0015670) pancreatic juice secretion(GO:0030157) saliva secretion(GO:0046541) |
| 0.0 |
0.5 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.1 |
GO:0015853 |
adenine transport(GO:0015853) |
| 0.0 |
0.1 |
GO:0006896 |
Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 |
0.1 |
GO:0003322 |
pancreatic A cell development(GO:0003322) |
| 0.0 |
0.1 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.1 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 |
0.0 |
GO:0060291 |
long-term synaptic potentiation(GO:0060291) |
| 0.0 |
0.0 |
GO:0046951 |
cellular ketone body metabolic process(GO:0046950) ketone body biosynthetic process(GO:0046951) |
| 0.0 |
0.0 |
GO:0042226 |
interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 |
0.1 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.0 |
0.1 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) chondrocyte development(GO:0002063) |
| 0.0 |
0.4 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 |
0.1 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 |
0.0 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.5 |
GO:0005980 |
glycogen catabolic process(GO:0005980) |
| 0.0 |
0.1 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.1 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 |
0.0 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.3 |
GO:0051156 |
glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 |
0.5 |
GO:0006699 |
bile acid biosynthetic process(GO:0006699) |
| 0.0 |
0.3 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.2 |
GO:0032091 |
negative regulation of protein binding(GO:0032091) |
| 0.0 |
0.1 |
GO:0018195 |
peptidyl-arginine modification(GO:0018195) |
| 0.0 |
0.0 |
GO:0045839 |
negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 |
0.0 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.0 |
GO:0042727 |
flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 |
0.1 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.0 |
0.1 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.0 |
0.0 |
GO:0006047 |
UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 |
0.1 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.0 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.0 |
0.1 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 |
0.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 |
0.8 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
0.1 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.0 |
0.0 |
GO:0003077 |
obsolete negative regulation of diuresis(GO:0003077) |
| 0.0 |
0.1 |
GO:0032096 |
negative regulation of response to food(GO:0032096) |
| 0.0 |
0.1 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 |
0.1 |
GO:0001575 |
globoside metabolic process(GO:0001575) |
| 0.0 |
0.0 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.0 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.0 |
0.1 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.1 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.1 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.0 |
0.0 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.0 |
0.1 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.2 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
0.1 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.0 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.2 |
GO:0001662 |
behavioral fear response(GO:0001662) |
| 0.0 |
0.0 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 |
0.1 |
GO:0050732 |
negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 |
0.0 |
GO:0006681 |
galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.0 |
0.2 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.0 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.0 |
GO:1901798 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 |
0.0 |
GO:0060603 |
mammary gland duct morphogenesis(GO:0060603) |
| 0.0 |
0.1 |
GO:0061036 |
positive regulation of cartilage development(GO:0061036) |
| 0.0 |
0.0 |
GO:0033522 |
histone H2A ubiquitination(GO:0033522) |
| 0.0 |
0.1 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.2 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 |
0.0 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 |
0.1 |
GO:0007141 |
male meiosis I(GO:0007141) |
| 0.0 |
0.2 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.0 |
0.1 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 |
0.1 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.0 |
GO:0006971 |
hypotonic response(GO:0006971) |
| 0.0 |
0.1 |
GO:0095500 |
G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.1 |
GO:0007632 |
visual behavior(GO:0007632) |
| 0.0 |
0.2 |
GO:0035050 |
embryonic heart tube development(GO:0035050) |
| 0.0 |
0.1 |
GO:0090090 |
negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 |
0.1 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.1 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.0 |
GO:0051974 |
negative regulation of telomerase activity(GO:0051974) |