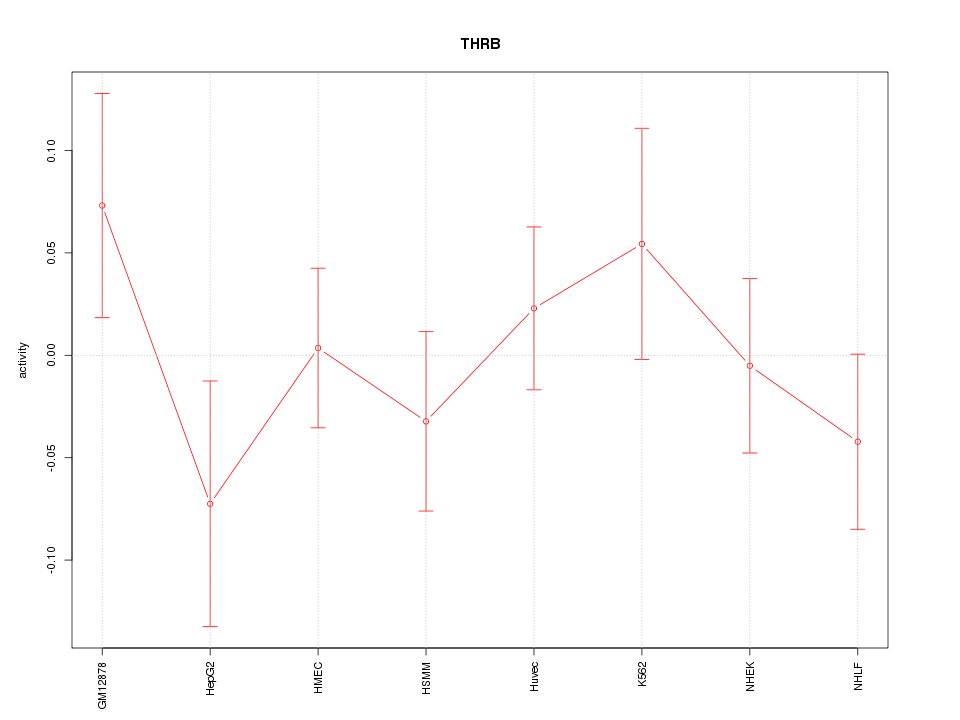
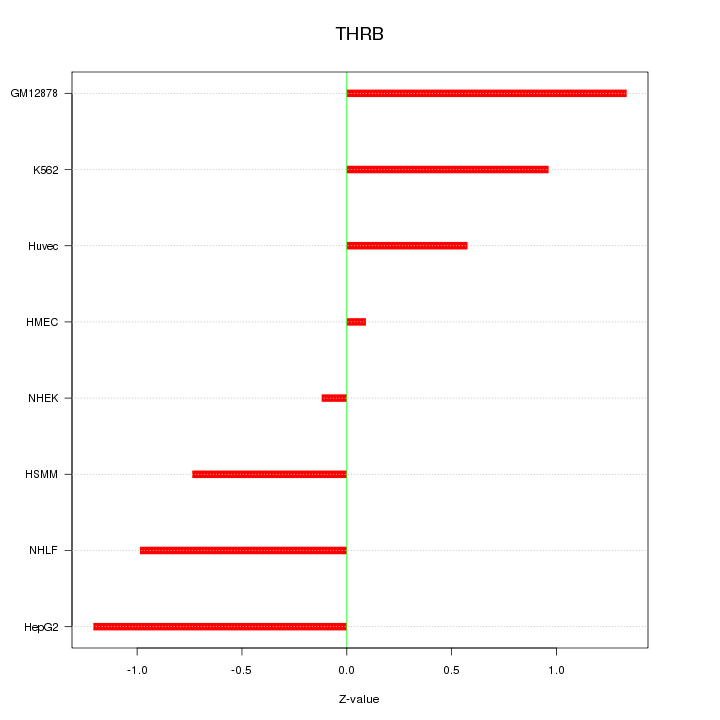
Motif ID: THRB
Z-value: 0.869
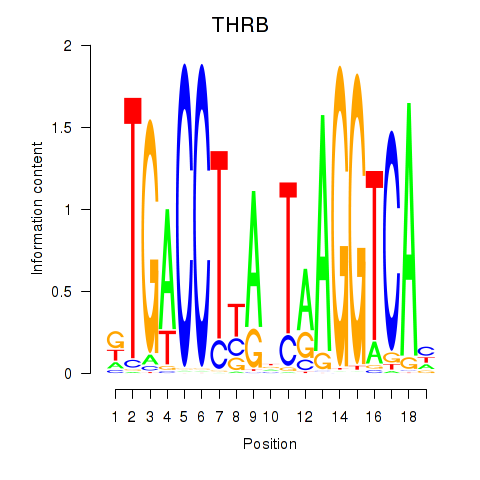
Transcription factors associated with THRB:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| THRB | ENSG00000151090.13 | THRB |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 1.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 0.9 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 2.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 0.5 | GO:0046732 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by virus of host immune response(GO:0075528) |
| 0.1 | 0.8 | GO:0032527 | retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.1 | 0.8 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 5.5 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 0.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.9 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.2 | 1.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 0.5 | GO:0015217 | purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.1 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |