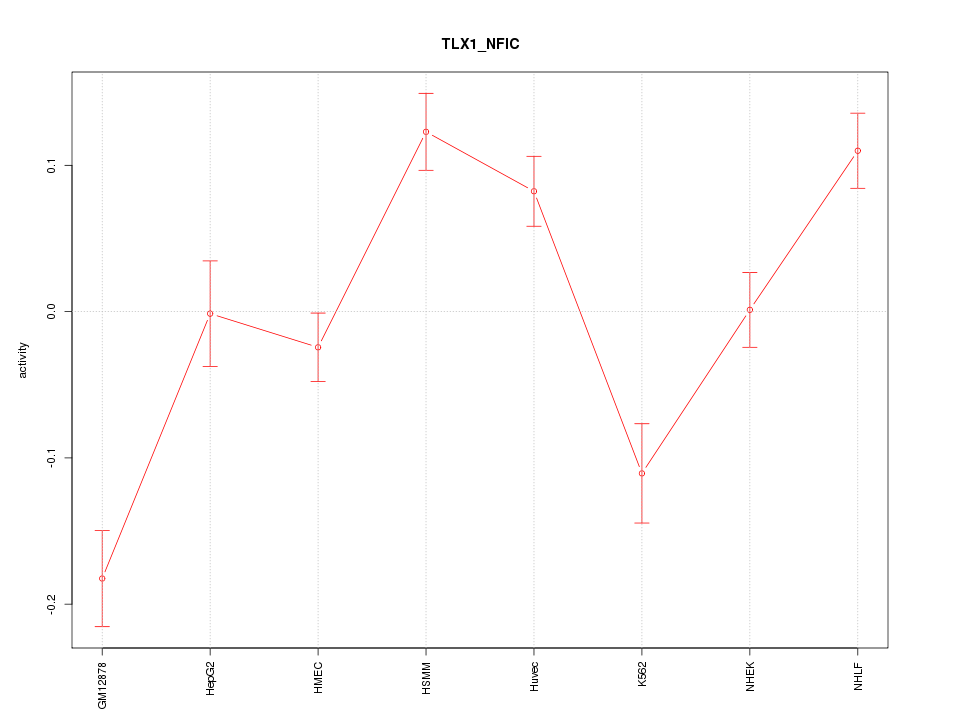
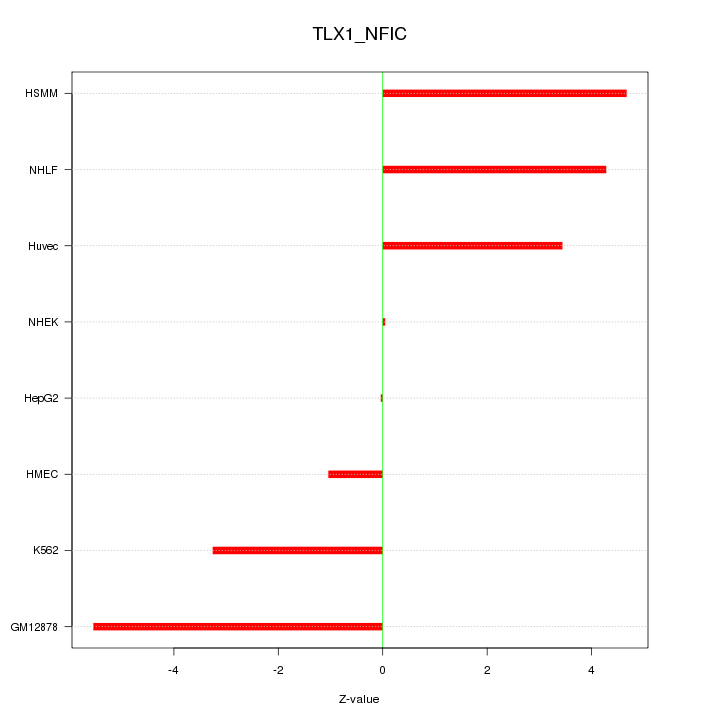
Motif ID: TLX1_NFIC
Z-value: 3.436
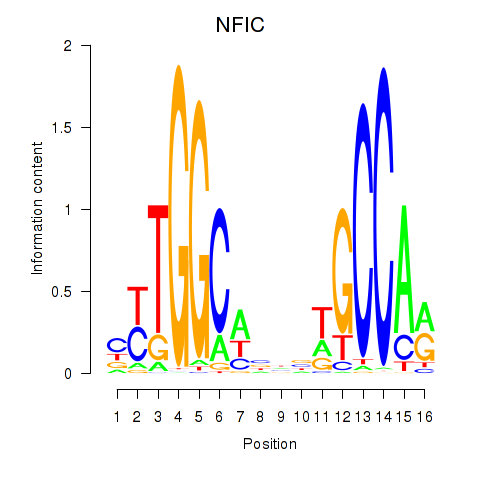
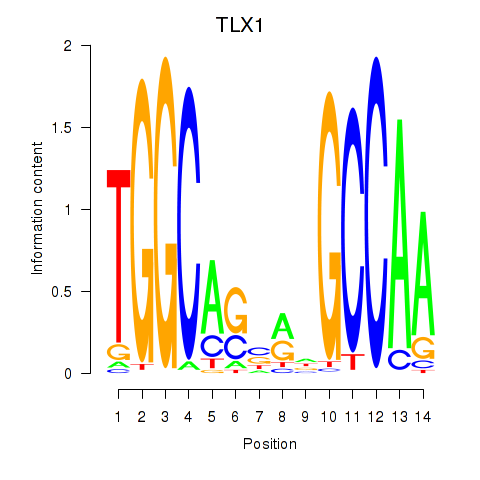
Transcription factors associated with TLX1_NFIC:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NFIC | ENSG00000141905.13 | NFIC |
| TLX1 | ENSG00000107807.8 | TLX1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 15.1 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 2.4 | 11.9 | GO:0044854 | membrane raft assembly(GO:0001765) nitric oxide homeostasis(GO:0033484) negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 2.2 | 6.6 | GO:0070483 | detection of hypoxia(GO:0070483) |
| 1.2 | 3.7 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 1.0 | 2.9 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 1.0 | 2.9 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.8 | 3.9 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.7 | 2.1 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.7 | 4.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.7 | 5.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.6 | 16.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.6 | 4.3 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.6 | 4.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.5 | 6.9 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.5 | 2.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.4 | 1.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.4 | 1.2 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.4 | 1.2 | GO:0031133 | regulation of axon diameter(GO:0031133) regulation of cell projection size(GO:0032536) |
| 0.4 | 3.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.3 | 3.4 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.3 | 2.3 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.3 | 7.7 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.3 | 3.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.3 | 0.8 | GO:0035120 | post-embryonic appendage morphogenesis(GO:0035120) |
| 0.3 | 2.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 2.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.3 | 2.4 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.3 | 2.8 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.3 | 1.0 | GO:0071071 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) regulation of phospholipid biosynthetic process(GO:0071071) |
| 0.2 | 3.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.2 | 2.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.6 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 6.5 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.2 | 3.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.5 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 1.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 0.6 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.1 | 2.5 | GO:0003407 | neural retina development(GO:0003407) |
| 0.1 | 3.1 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 9.9 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.1 | 0.5 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 1.1 | GO:0010875 | positive regulation of cholesterol efflux(GO:0010875) |
| 0.1 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.8 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.9 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.1 | 14.9 | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043281) |
| 0.1 | 6.4 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 4.5 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.1 | 0.3 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.1 | 0.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 6.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 2.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 3.5 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.1 | 0.7 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.3 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.5 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.1 | 0.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 5.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 2.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.5 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 1.8 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 | 0.5 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 2.5 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 3.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.5 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 1.7 | GO:0051318 | G1 phase(GO:0051318) |
| 0.0 | 13.7 | GO:0016049 | cell growth(GO:0016049) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 7.8 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 2.1 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.1 | GO:0014888 | striated muscle adaptation(GO:0014888) |
| 0.0 | 2.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 1.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.6 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.3 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 1.5 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) |
| 0.0 | 1.0 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.3 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) syncytium formation(GO:0006949) myoblast fusion(GO:0007520) |
| 0.0 | 0.4 | GO:0051181 | cofactor transport(GO:0051181) sulfur compound transport(GO:0072348) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.9 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.2 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.1 | 4.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.9 | 3.5 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.9 | 14.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.7 | 2.8 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.6 | 7.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 3.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 6.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.4 | 11.5 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.4 | 3.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.3 | 3.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 3.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 19.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.2 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 0.5 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 1.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 5.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.3 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.1 | 0.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 4.9 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 6.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 36.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 6.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 6.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 2.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.9 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 12.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 1.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.6 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 6.8 | GO:0005624 | obsolete membrane fraction(GO:0005624) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0005534 | galactose binding(GO:0005534) |
| 0.9 | 15.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.9 | 6.4 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.9 | 11.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.9 | 5.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.9 | 4.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.8 | 4.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 5.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.8 | 3.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.6 | 2.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.4 | 2.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.4 | 10.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 1.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 2.8 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 3.7 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.3 | 3.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 1.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 2.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 5.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 3.4 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.2 | 14.3 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 1.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 1.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 3.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 2.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 0.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 2.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.2 | 3.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 2.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 0.6 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 0.6 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.2 | 2.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.6 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 23.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 4.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 2.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.1 | 0.5 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 1.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 8.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 8.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 0.8 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 2.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 5.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.9 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.1 | 1.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 6.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 3.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 2.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.7 | GO:0016462 | pyrophosphatase activity(GO:0016462) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 5.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 27.0 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.2 | 2.8 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.1 | 0.3 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.0 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.1 | 1.5 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.5 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.8 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.5 | ST_GA13_PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.2 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |