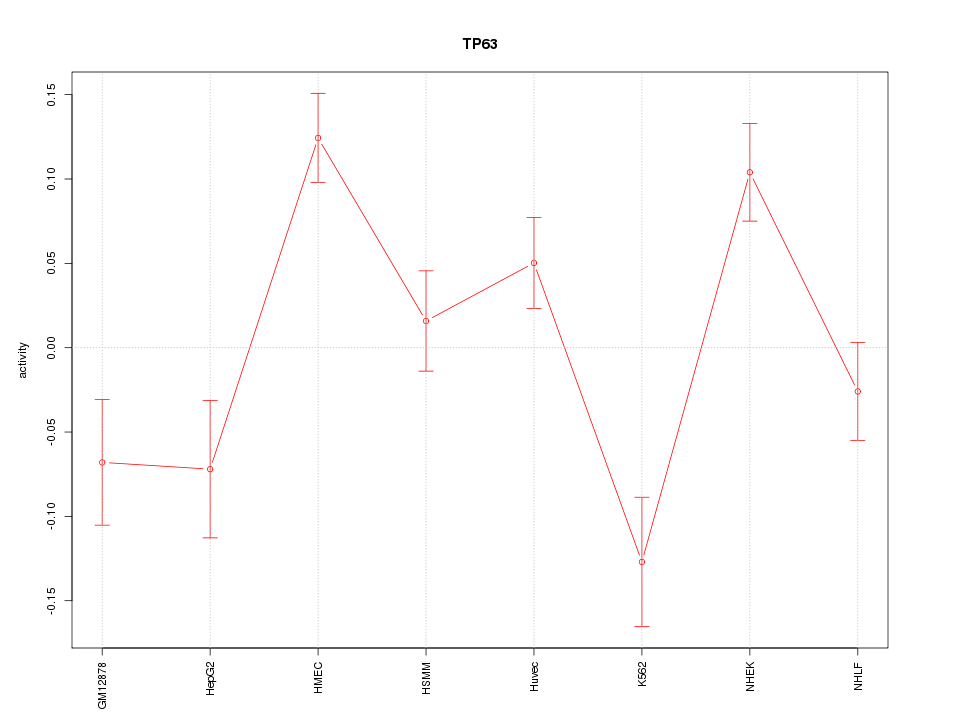
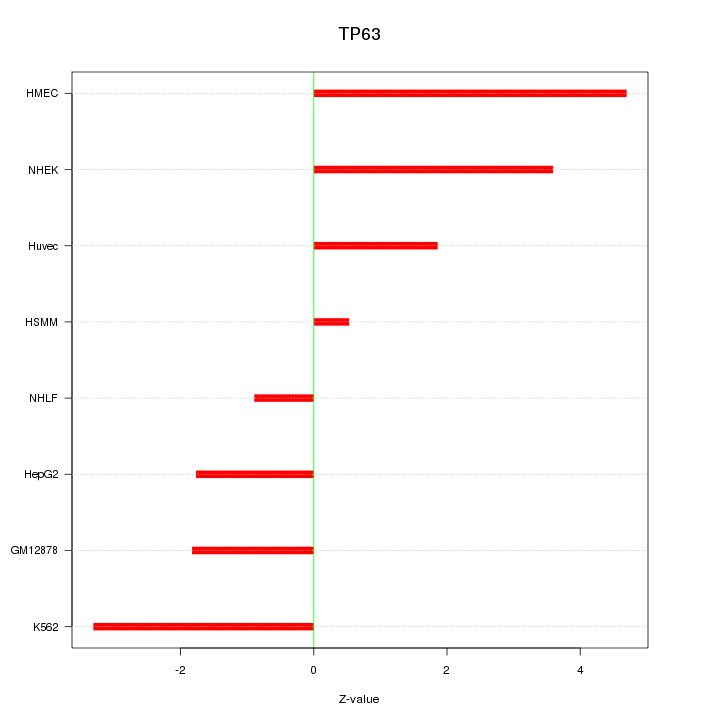
Motif ID: TP63
Z-value: 2.668
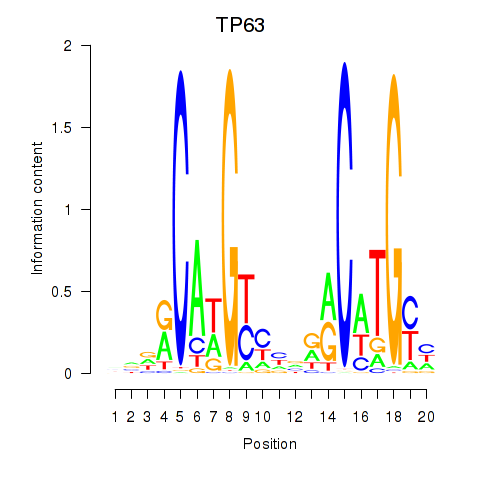
Transcription factors associated with TP63:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| TP63 | ENSG00000073282.8 | TP63 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 1.8 | 7.1 | GO:0051503 | purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 1.2 | 5.0 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.7 | 5.0 | GO:0090025 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.6 | 2.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.5 | 3.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.5 | 2.3 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.4 | 12.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.4 | 3.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.3 | 0.7 | GO:0021873 | forebrain neuroblast division(GO:0021873) neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.3 | 3.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 3.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.3 | 1.5 | GO:1903008 | organelle disassembly(GO:1903008) |
| 0.3 | 8.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 4.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 1.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.3 | 8.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 1.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.2 | 1.7 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.2 | 0.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 0.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.2 | 3.8 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.2 | 1.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 0.9 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.2 | 2.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 6.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 6.0 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 10.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 2.0 | GO:1901224 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 0.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 3.4 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 4.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0016559 | peroxisome membrane biogenesis(GO:0016557) peroxisome fission(GO:0016559) |
| 0.0 | 1.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 5.5 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.3 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.2 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.3 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.2 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 5.0 | GO:0006184 | obsolete GTP catabolic process(GO:0006184) |
| 0.0 | 0.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.2 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.4 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.6 | 7.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 2.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.3 | 13.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 4.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 1.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 6.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 1.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.7 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.2 | 2.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 4.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 3.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 4.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 3.4 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 1.9 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 5.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 3.3 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.0 | 11.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.9 | GO:0030054 | cell junction(GO:0030054) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 7.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.6 | 5.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 4.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 0.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 2.7 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.3 | 1.7 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.3 | 5.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 3.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 3.7 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 11.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.9 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 1.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 11.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 5.0 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.1 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 1.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 9.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 4.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 9.1 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.9 | GO:0016566 | obsolete specific transcriptional repressor activity(GO:0016566) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 7.9 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 4.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 2.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 4.4 | GO:0032403 | protein complex binding(GO:0032403) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |