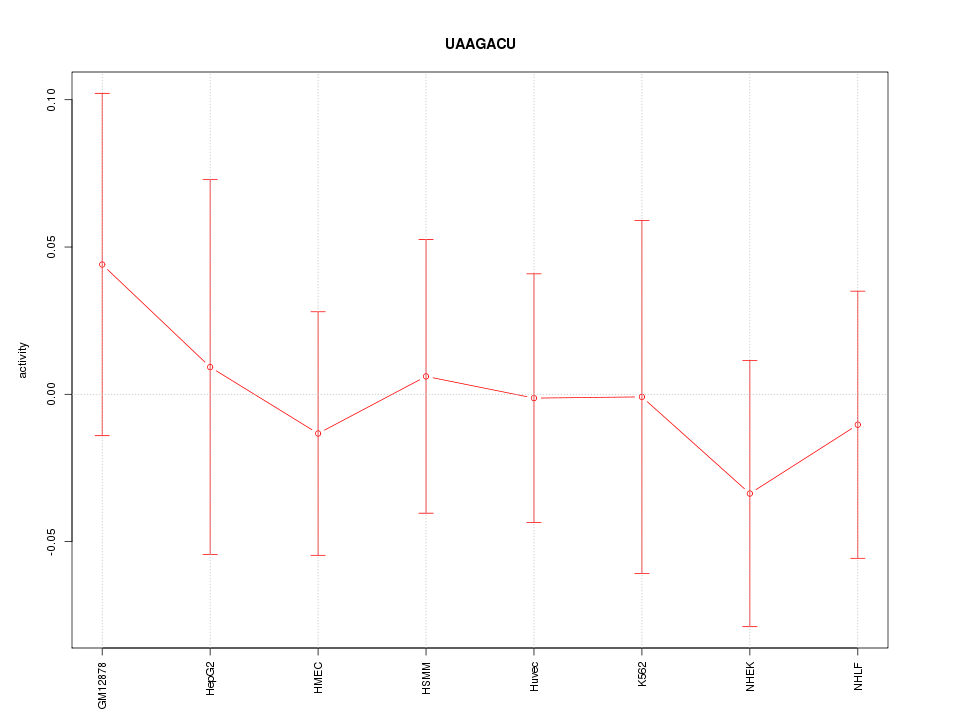
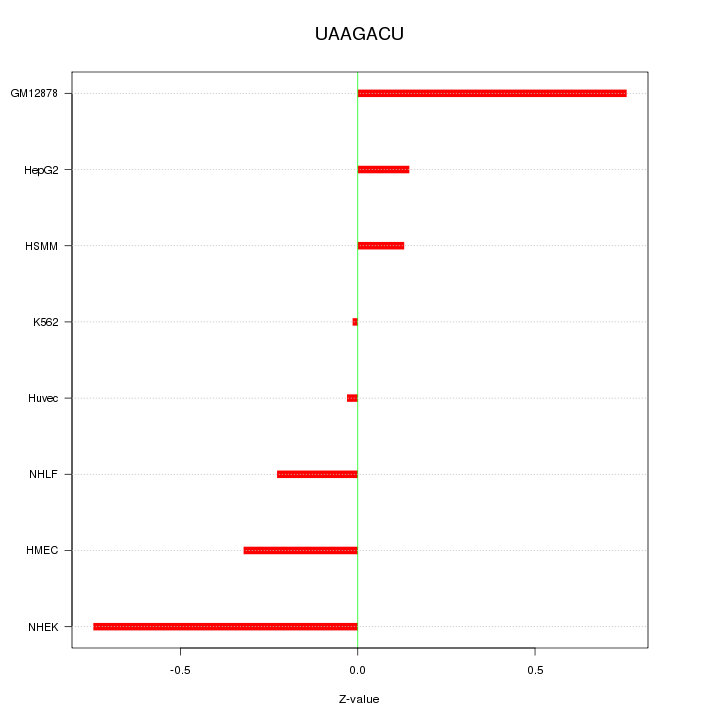
|
chr8_+_38614807
|
0.460
|
ENST00000330691.6
ENST00000348567.4
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr19_-_7293942
|
0.259
|
ENST00000341500.5
ENST00000302850.5
|
INSR
|
insulin receptor
|
|
chr1_-_154842741
|
0.208
|
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr8_+_56014949
|
0.207
|
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr10_+_112631547
|
0.189
|
ENST00000280154.7
ENST00000393104.2
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr7_-_120498357
|
0.184
|
ENST00000415871.1
ENST00000222747.3
ENST00000430985.1
|
TSPAN12
|
tetraspanin 12
|
|
chr5_+_112312416
|
0.171
|
ENST00000389063.2
|
DCP2
|
decapping mRNA 2
|
|
chr17_+_58677539
|
0.168
|
ENST00000305921.3
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr2_-_64881018
|
0.166
|
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2
|
|
chr3_+_186501336
|
0.162
|
ENST00000323963.5
ENST00000440191.2
ENST00000356531.5
|
EIF4A2
|
eukaryotic translation initiation factor 4A2
|
|
chr6_-_99797522
|
0.149
|
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila)
|
|
chr16_-_46655538
|
0.137
|
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chrY_+_15016725
|
0.136
|
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked
|
|
chr3_-_72496035
|
0.107
|
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein
|
|
chr7_-_139876812
|
0.105
|
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A
|
|
chr10_+_63661053
|
0.102
|
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr3_+_43328004
|
0.082
|
ENST00000454177.1
ENST00000429705.2
ENST00000296088.7
ENST00000437827.1
|
SNRK
|
SNF related kinase
|
|
chr5_-_76788317
|
0.081
|
ENST00000296679.4
|
WDR41
|
WD repeat domain 41
|
|
chr1_+_145611010
|
0.080
|
ENST00000369291.5
|
RNF115
|
ring finger protein 115
|
|
chr1_-_16678914
|
0.078
|
ENST00000375592.3
|
FBXO42
|
F-box protein 42
|
|
chr2_+_228029281
|
0.077
|
ENST00000396578.3
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen)
|
|
chr2_-_86564776
|
0.076
|
ENST00000165698.5
ENST00000541910.1
ENST00000535845.1
|
REEP1
|
receptor accessory protein 1
|
|
chr8_+_58907104
|
0.075
|
ENST00000361488.3
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr12_+_72148614
|
0.074
|
ENST00000261263.3
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr17_-_60142609
|
0.074
|
ENST00000397786.2
|
MED13
|
mediator complex subunit 13
|
|
chr3_-_15374033
|
0.068
|
ENST00000253688.5
ENST00000383791.3
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr2_+_42396472
|
0.067
|
ENST00000318522.5
ENST00000402711.2
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr15_-_49338748
|
0.065
|
ENST00000559471.1
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr10_-_27149792
|
0.063
|
ENST00000376140.3
ENST00000376170.4
|
ABI1
|
abl-interactor 1
|
|
chr11_-_86666427
|
0.062
|
ENST00000531380.1
|
FZD4
|
frizzled family receptor 4
|
|
chr12_+_68042495
|
0.061
|
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2
|
|
chr7_+_90225796
|
0.061
|
ENST00000380050.3
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr12_-_12419703
|
0.060
|
ENST00000543091.1
ENST00000261349.4
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr1_+_97187318
|
0.057
|
ENST00000609116.1
ENST00000370198.1
ENST00000370197.1
ENST00000426398.2
ENST00000394184.3
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr8_+_42752053
|
0.056
|
ENST00000307602.4
|
HOOK3
|
hook microtubule-tethering protein 3
|
|
chr2_-_68290106
|
0.052
|
ENST00000407324.1
ENST00000355848.3
ENST00000409302.1
ENST00000410067.3
|
C1D
|
C1D nuclear receptor corepressor
|
|
chr6_+_72596604
|
0.051
|
ENST00000348717.5
ENST00000517960.1
ENST00000518273.1
ENST00000522291.1
ENST00000521978.1
ENST00000520567.1
ENST00000264839.7
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr17_+_29718642
|
0.050
|
ENST00000325874.8
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr14_-_50698276
|
0.049
|
ENST00000216373.5
|
SOS2
|
son of sevenless homolog 2 (Drosophila)
|
|
chr8_+_11561660
|
0.049
|
ENST00000526716.1
ENST00000335135.4
ENST00000528027.1
|
GATA4
|
GATA binding protein 4
|
|
chr17_+_66508537
|
0.049
|
ENST00000392711.1
ENST00000585427.1
ENST00000589228.1
ENST00000536854.2
ENST00000588702.1
ENST00000589309.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha
|
|
chr1_+_36396677
|
0.049
|
ENST00000373191.4
ENST00000397828.2
|
AGO3
|
argonaute RISC catalytic component 3
|
|
chr22_+_21771656
|
0.048
|
ENST00000407464.2
|
HIC2
|
hypermethylated in cancer 2
|
|
chr1_-_39339777
|
0.048
|
ENST00000397572.2
|
MYCBP
|
MYC binding protein
|
|
chr5_+_161274685
|
0.047
|
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_122110691
|
0.045
|
ENST00000379516.2
ENST00000505934.1
ENST00000514949.1
|
SNX2
|
sorting nexin 2
|
|
chr2_+_120517174
|
0.045
|
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte)
|
|
chr15_+_57210818
|
0.044
|
ENST00000438423.2
ENST00000267811.5
ENST00000452095.2
ENST00000559609.1
ENST00000333725.5
|
TCF12
|
transcription factor 12
|
|
chr15_-_64648273
|
0.041
|
ENST00000607537.1
ENST00000303052.7
ENST00000303032.6
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr1_+_50574585
|
0.041
|
ENST00000371824.1
ENST00000371823.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4
|
|
chr6_-_13711773
|
0.038
|
ENST00000011619.3
|
RANBP9
|
RAN binding protein 9
|
|
chr3_+_29322803
|
0.037
|
ENST00000396583.3
ENST00000383767.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr17_-_45266542
|
0.037
|
ENST00000531206.1
ENST00000527547.1
ENST00000446365.2
ENST00000575483.1
ENST00000066544.3
|
CDC27
|
cell division cycle 27
|
|
chrX_+_16737718
|
0.037
|
ENST00000380155.3
|
SYAP1
|
synapse associated protein 1
|
|
chr5_-_114880533
|
0.032
|
ENST00000274457.3
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chrX_+_41192595
|
0.032
|
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked
|
|
chr11_-_128392085
|
0.029
|
ENST00000526145.2
ENST00000531611.1
ENST00000319397.6
ENST00000345075.4
ENST00000535549.1
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1
|
|
chr12_-_42538657
|
0.028
|
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr9_-_19786926
|
0.028
|
ENST00000341998.2
ENST00000286344.3
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr6_-_166075557
|
0.027
|
ENST00000539869.2
ENST00000366882.1
|
PDE10A
|
phosphodiesterase 10A
|
|
chr15_+_101142722
|
0.025
|
ENST00000332783.7
ENST00000558747.1
ENST00000343276.4
|
ASB7
|
ankyrin repeat and SOCS box containing 7
|
|
chr15_+_44829255
|
0.024
|
ENST00000261868.5
ENST00000424492.3
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr17_-_48785216
|
0.024
|
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr7_+_65338230
|
0.023
|
ENST00000360768.3
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1
|
|
chr1_-_39347255
|
0.021
|
ENST00000454994.2
ENST00000357771.3
|
GJA9
|
gap junction protein, alpha 9, 59kDa
|
|
chr2_+_176981307
|
0.021
|
ENST00000249501.4
|
HOXD10
|
homeobox D10
|
|
chr13_+_49550015
|
0.021
|
ENST00000492622.2
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr9_-_23821273
|
0.020
|
ENST00000380110.4
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2
|
|
chr7_+_138916231
|
0.019
|
ENST00000473989.3
ENST00000288561.8
|
UBN2
|
ubinuclein 2
|
|
chr1_-_21113105
|
0.018
|
ENST00000375000.1
ENST00000419490.1
ENST00000414993.1
ENST00000443615.1
ENST00000312239.5
|
HP1BP3
|
heterochromatin protein 1, binding protein 3
|
|
chr6_+_163835669
|
0.018
|
ENST00000453779.2
ENST00000275262.7
ENST00000392127.2
ENST00000361752.3
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr13_+_102104980
|
0.018
|
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr14_+_105781048
|
0.018
|
ENST00000458164.2
ENST00000447393.1
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr7_-_44924939
|
0.017
|
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B
|
|
chr6_+_133562472
|
0.016
|
ENST00000430974.2
ENST00000367895.5
ENST00000355167.3
ENST00000355286.6
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr4_+_139936905
|
0.015
|
ENST00000280614.2
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae)
|
|
chr3_-_15563229
|
0.014
|
ENST00000383786.5
ENST00000383787.2
ENST00000383785.2
ENST00000383788.5
ENST00000603808.1
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr9_+_82186872
|
0.012
|
ENST00000376544.3
ENST00000376520.4
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr13_+_50571155
|
0.010
|
ENST00000420995.2
ENST00000378182.3
ENST00000356017.4
ENST00000457662.2
|
TRIM13
|
tripartite motif containing 13
|
|
chr8_-_89339705
|
0.008
|
ENST00000286614.6
|
MMP16
|
matrix metallopeptidase 16 (membrane-inserted)
|
|
chr3_+_89156674
|
0.006
|
ENST00000336596.2
|
EPHA3
|
EPH receptor A3
|
|
chr1_+_43148059
|
0.006
|
ENST00000321358.7
ENST00000332220.6
|
YBX1
|
Y box binding protein 1
|
|
chr6_+_37225540
|
0.005
|
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr2_+_176987088
|
0.005
|
ENST00000249499.6
|
HOXD9
|
homeobox D9
|
|
chr5_-_79287060
|
0.004
|
ENST00000512560.1
ENST00000509852.1
ENST00000512528.1
|
MTX3
|
metaxin 3
|
|
chr4_+_123747834
|
0.004
|
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr2_-_131850951
|
0.002
|
ENST00000409185.1
ENST00000389915.3
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr21_-_16437255
|
0.002
|
ENST00000400199.1
ENST00000400202.1
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr15_-_56535464
|
0.002
|
ENST00000559447.2
ENST00000422057.1
ENST00000317318.6
ENST00000423270.1
|
RFX7
|
regulatory factor X, 7
|
|
chr14_-_61116168
|
0.002
|
ENST00000247182.6
|
SIX1
|
SIX homeobox 1
|