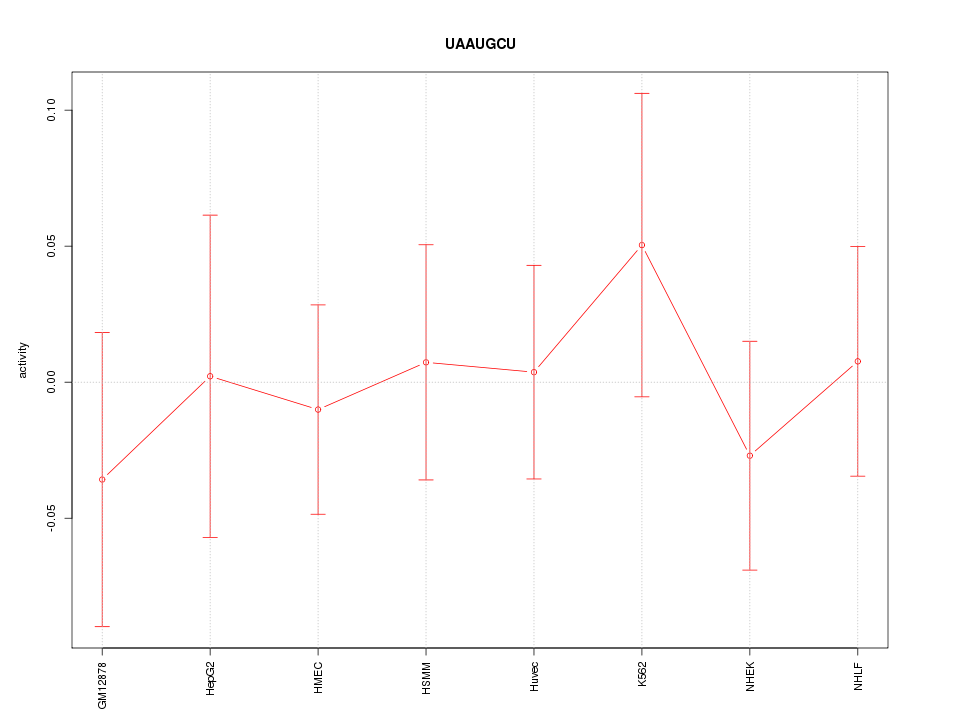
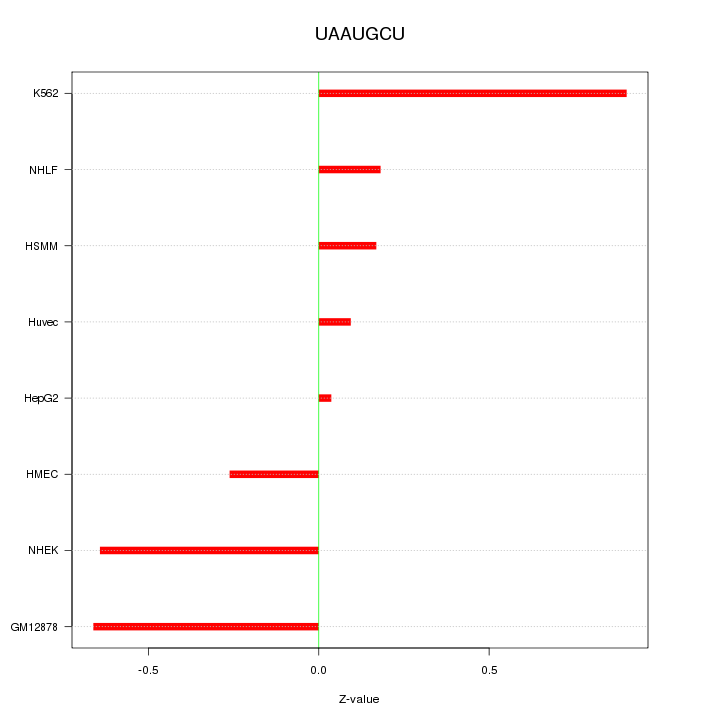
|
chr17_-_13505219
|
0.541
|
ENST00000284110.1
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr11_-_85780086
|
0.362
|
ENST00000532317.1
ENST00000528256.1
ENST00000526033.1
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr2_+_102508955
|
0.325
|
ENST00000414004.2
|
FLJ20373
|
FLJ20373
|
|
chr15_+_96873921
|
0.314
|
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr6_+_135502466
|
0.306
|
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog
|
|
chr14_-_30396948
|
0.289
|
ENST00000331968.5
|
PRKD1
|
protein kinase D1
|
|
chr5_+_112312416
|
0.285
|
ENST00000389063.2
|
DCP2
|
decapping mRNA 2
|
|
chr11_+_35684288
|
0.239
|
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44
|
|
chr2_-_208634287
|
0.221
|
ENST00000295417.3
|
FZD5
|
frizzled family receptor 5
|
|
chr20_+_48807351
|
0.221
|
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr2_-_200322723
|
0.208
|
ENST00000417098.1
|
SATB2
|
SATB homeobox 2
|
|
chr1_+_114522049
|
0.197
|
ENST00000369551.1
ENST00000320334.4
|
OLFML3
|
olfactomedin-like 3
|
|
chr8_-_116681221
|
0.184
|
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr7_+_66093851
|
0.181
|
ENST00000275532.3
|
KCTD7
|
potassium channel tetramerization domain containing 7
|
|
chr21_+_30671189
|
0.170
|
ENST00000286800.3
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr2_+_70056762
|
0.165
|
ENST00000282570.3
|
GMCL1
|
germ cell-less, spermatogenesis associated 1
|
|
chr2_+_48010221
|
0.159
|
ENST00000234420.5
|
MSH6
|
mutS homolog 6
|
|
chr3_-_141868357
|
0.158
|
ENST00000489671.1
ENST00000475734.1
ENST00000467072.1
ENST00000499676.2
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr14_+_103058948
|
0.158
|
ENST00000262241.6
|
RCOR1
|
REST corepressor 1
|
|
chr8_-_105601134
|
0.150
|
ENST00000276654.5
ENST00000424843.2
|
LRP12
|
low density lipoprotein receptor-related protein 12
|
|
chr9_-_16870704
|
0.150
|
ENST00000380672.4
ENST00000380667.2
ENST00000380666.2
ENST00000486514.1
|
BNC2
|
basonuclin 2
|
|
chr1_+_215740709
|
0.143
|
ENST00000259154.4
|
KCTD3
|
potassium channel tetramerization domain containing 3
|
|
chr7_-_151217001
|
0.132
|
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain
|
|
chr11_-_46867780
|
0.129
|
ENST00000529230.1
ENST00000415402.1
ENST00000312055.5
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr12_-_25102252
|
0.129
|
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr5_+_135468516
|
0.123
|
ENST00000507118.1
ENST00000511116.1
ENST00000545279.1
ENST00000545620.1
|
SMAD5
|
SMAD family member 5
|
|
chr17_-_4167142
|
0.118
|
ENST00000570535.1
ENST00000574367.1
ENST00000341657.4
ENST00000433651.1
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr1_+_63833261
|
0.116
|
ENST00000371108.4
|
ALG6
|
ALG6, alpha-1,3-glucosyltransferase
|
|
chr11_+_47279504
|
0.112
|
ENST00000441012.2
ENST00000437276.1
ENST00000436029.1
ENST00000467728.1
ENST00000405853.3
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr2_+_28615669
|
0.112
|
ENST00000379619.1
ENST00000264716.4
|
FOSL2
|
FOS-like antigen 2
|
|
chr8_+_104033277
|
0.111
|
ENST00000518857.1
ENST00000395862.3
|
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1
|
|
chr1_-_229569834
|
0.108
|
ENST00000366684.3
ENST00000366683.2
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr22_-_38380543
|
0.106
|
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chr2_+_208394616
|
0.102
|
ENST00000432329.2
ENST00000353267.3
ENST00000445803.1
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr1_+_36396677
|
0.098
|
ENST00000373191.4
ENST00000397828.2
|
AGO3
|
argonaute RISC catalytic component 3
|
|
chr12_+_72233487
|
0.098
|
ENST00000482439.2
ENST00000550746.1
ENST00000491063.1
ENST00000319106.8
ENST00000485960.2
ENST00000393309.3
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr1_+_2160134
|
0.096
|
ENST00000378536.4
|
SKI
|
v-ski avian sarcoma viral oncogene homolog
|
|
chr3_-_123603137
|
0.095
|
ENST00000360304.3
ENST00000359169.1
ENST00000346322.5
ENST00000360772.3
|
MYLK
|
myosin light chain kinase
|
|
chr7_-_32931387
|
0.094
|
ENST00000304056.4
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr7_-_139876812
|
0.094
|
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A
|
|
chr2_+_46926048
|
0.094
|
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr3_-_160283348
|
0.091
|
ENST00000334256.4
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr4_+_123747834
|
0.090
|
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr1_-_70671216
|
0.087
|
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr10_+_69644404
|
0.085
|
ENST00000212015.6
|
SIRT1
|
sirtuin 1
|
|
chrX_-_20284958
|
0.084
|
ENST00000379565.3
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr14_+_75536280
|
0.079
|
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C
|
|
chr12_+_69004619
|
0.078
|
ENST00000250559.9
ENST00000393436.5
ENST00000425247.2
ENST00000489473.2
ENST00000422358.2
ENST00000541167.1
ENST00000538283.1
ENST00000341355.5
ENST00000537460.1
ENST00000450214.2
ENST00000545270.1
ENST00000538980.1
ENST00000542018.1
ENST00000543393.1
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr9_-_132597529
|
0.078
|
ENST00000372447.3
|
C9orf78
|
chromosome 9 open reading frame 78
|
|
chr4_+_41937131
|
0.078
|
ENST00000504986.1
ENST00000508448.1
ENST00000513702.1
ENST00000325094.5
|
TMEM33
|
transmembrane protein 33
|
|
chr1_-_235292250
|
0.077
|
ENST00000366607.4
|
TOMM20
|
translocase of outer mitochondrial membrane 20 homolog (yeast)
|
|
chr11_+_34642656
|
0.077
|
ENST00000257831.3
ENST00000450654.2
|
EHF
|
ets homologous factor
|
|
chr15_+_89631381
|
0.076
|
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr5_+_54603566
|
0.076
|
ENST00000230640.5
|
SKIV2L2
|
superkiller viralicidic activity 2-like 2 (S. cerevisiae)
|
|
chr17_+_57642886
|
0.075
|
ENST00000251241.4
ENST00000451169.2
ENST00000425628.3
ENST00000584385.1
ENST00000580030.1
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr12_+_93771659
|
0.073
|
ENST00000337179.5
ENST00000415493.2
|
NUDT4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
|
|
chr17_-_40306934
|
0.072
|
ENST00000550406.1
ENST00000547517.1
ENST00000393860.3
ENST00000346213.4
ENST00000592574.1
|
RAB5C
CTD-2132N18.3
|
RAB5C, member RAS oncogene family
Uncharacterized protein
|
|
chr21_-_34144157
|
0.071
|
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1
|
|
chr3_+_32147997
|
0.071
|
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr17_-_65241281
|
0.070
|
ENST00000358691.5
ENST00000580168.1
|
HELZ
|
helicase with zinc finger
|
|
chr7_-_14029515
|
0.065
|
ENST00000430479.1
ENST00000405218.2
ENST00000343495.5
|
ETV1
|
ets variant 1
|
|
chr6_+_7107999
|
0.065
|
ENST00000491191.1
ENST00000379938.2
ENST00000471433.1
|
RREB1
|
ras responsive element binding protein 1
|
|
chr5_-_9546180
|
0.065
|
ENST00000382496.5
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr16_-_20911641
|
0.064
|
ENST00000324344.4
ENST00000564349.1
|
DCUN1D3
ERI2
|
DCN1, defective in cullin neddylation 1, domain containing 3
ERI1 exoribonuclease family member 2
|
|
chr5_+_141488070
|
0.064
|
ENST00000253814.4
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr5_-_160973649
|
0.064
|
ENST00000393959.1
ENST00000517547.1
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr18_+_67956135
|
0.063
|
ENST00000397942.3
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr2_+_5832799
|
0.061
|
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr11_-_70507901
|
0.060
|
ENST00000449833.2
ENST00000357171.3
ENST00000449116.2
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr4_+_38665810
|
0.060
|
ENST00000261438.5
ENST00000514033.1
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr3_+_178866199
|
0.058
|
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha
|
|
chr15_-_89089860
|
0.054
|
ENST00000558413.1
ENST00000564406.1
ENST00000268148.8
|
DET1
|
de-etiolated homolog 1 (Arabidopsis)
|
|
chr16_+_67063036
|
0.054
|
ENST00000290858.6
ENST00000564034.1
|
CBFB
|
core-binding factor, beta subunit
|
|
chr3_-_113465065
|
0.053
|
ENST00000497255.1
ENST00000478020.1
ENST00000240922.3
ENST00000493900.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr18_-_53255766
|
0.053
|
ENST00000566286.1
ENST00000564999.1
ENST00000566279.1
ENST00000354452.3
ENST00000356073.4
|
TCF4
|
transcription factor 4
|
|
chr12_-_15038779
|
0.051
|
ENST00000228938.5
ENST00000539261.1
|
MGP
|
matrix Gla protein
|
|
chr5_+_112043186
|
0.051
|
ENST00000509732.1
ENST00000457016.1
ENST00000507379.1
|
APC
|
adenomatous polyposis coli
|
|
chr20_+_11871371
|
0.050
|
ENST00000254977.3
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr2_-_174830430
|
0.049
|
ENST00000310015.6
ENST00000455789.2
|
SP3
|
Sp3 transcription factor
|
|
chr21_-_40685477
|
0.048
|
ENST00000342449.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1
|
|
chr15_+_77223960
|
0.047
|
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain
|
|
chr17_-_48474828
|
0.047
|
ENST00000576448.1
ENST00000225972.7
|
LRRC59
|
leucine rich repeat containing 59
|
|
chr1_+_193091080
|
0.046
|
ENST00000367435.3
|
CDC73
|
cell division cycle 73
|
|
chr12_-_54582655
|
0.046
|
ENST00000504338.1
ENST00000514685.1
ENST00000504797.1
ENST00000513838.1
ENST00000505128.1
ENST00000337581.3
ENST00000503306.1
ENST00000243112.5
ENST00000514196.1
ENST00000506169.1
ENST00000507904.1
ENST00000508394.2
|
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1
|
|
chr15_+_50716576
|
0.045
|
ENST00000560297.1
ENST00000307179.4
ENST00000396444.3
ENST00000433963.1
ENST00000425032.3
|
USP8
|
ubiquitin specific peptidase 8
|
|
chr17_-_31204124
|
0.044
|
ENST00000579584.1
ENST00000318217.5
ENST00000583621.1
|
MYO1D
|
myosin ID
|
|
chrX_+_49687216
|
0.044
|
ENST00000376088.3
|
CLCN5
|
chloride channel, voltage-sensitive 5
|
|
chr2_-_64881018
|
0.044
|
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2
|
|
chr8_+_73921085
|
0.042
|
ENST00000276603.5
ENST00000276602.6
ENST00000518874.1
|
TERF1
|
telomeric repeat binding factor (NIMA-interacting) 1
|
|
chr3_+_180630090
|
0.040
|
ENST00000357559.4
ENST00000305586.7
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr12_+_104458235
|
0.039
|
ENST00000229330.4
|
HCFC2
|
host cell factor C2
|
|
chr12_-_57400227
|
0.038
|
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr7_+_94139105
|
0.038
|
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1
|
|
chr16_-_11350036
|
0.037
|
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr2_-_24149977
|
0.036
|
ENST00000238789.5
|
ATAD2B
|
ATPase family, AAA domain containing 2B
|
|
chr8_+_40010989
|
0.035
|
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr6_+_108881012
|
0.035
|
ENST00000343882.6
|
FOXO3
|
forkhead box O3
|
|
chr1_-_46216286
|
0.034
|
ENST00000396478.3
ENST00000359942.4
|
IPP
|
intracisternal A particle-promoted polypeptide
|
|
chr7_+_106809406
|
0.034
|
ENST00000468410.1
ENST00000478930.1
ENST00000464009.1
ENST00000222574.4
|
HBP1
|
HMG-box transcription factor 1
|
|
chr12_-_100536608
|
0.033
|
ENST00000356828.3
ENST00000279907.7
|
UHRF1BP1L
|
UHRF1 binding protein 1-like
|
|
chr7_+_155250824
|
0.033
|
ENST00000297375.4
|
EN2
|
engrailed homeobox 2
|
|
chrX_-_54384425
|
0.032
|
ENST00000375169.3
ENST00000354646.2
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr19_+_4402659
|
0.029
|
ENST00000301280.5
ENST00000585854.1
|
CHAF1A
|
chromatin assembly factor 1, subunit A (p150)
|
|
chr9_-_14314066
|
0.029
|
ENST00000397575.3
|
NFIB
|
nuclear factor I/B
|
|
chr22_-_50217981
|
0.028
|
ENST00000457780.2
|
BRD1
|
bromodomain containing 1
|
|
chr4_-_76598296
|
0.026
|
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr10_-_13390270
|
0.025
|
ENST00000378614.4
ENST00000545675.1
ENST00000327347.5
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr13_-_29069232
|
0.023
|
ENST00000282397.4
ENST00000541932.1
ENST00000539099.1
|
FLT1
|
fms-related tyrosine kinase 1
|
|
chr21_+_38071430
|
0.023
|
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2
|
|
chr14_-_34931458
|
0.022
|
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A
|
|
chr7_-_155604967
|
0.022
|
ENST00000297261.2
|
SHH
|
sonic hedgehog
|
|
chrX_+_122993827
|
0.022
|
ENST00000371199.3
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr17_+_35849937
|
0.021
|
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14
|
|
chr3_-_18466787
|
0.021
|
ENST00000338745.6
ENST00000450898.1
|
SATB1
|
SATB homeobox 1
|
|
chr1_-_52456352
|
0.021
|
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr10_-_112064665
|
0.021
|
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr1_+_214161272
|
0.020
|
ENST00000498508.2
ENST00000366958.4
|
PROX1
|
prospero homeobox 1
|
|
chr5_-_149492904
|
0.019
|
ENST00000286301.3
ENST00000511344.1
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr9_+_91003271
|
0.019
|
ENST00000375859.3
ENST00000541629.1
|
SPIN1
|
spindlin 1
|
|
chr8_-_23540402
|
0.018
|
ENST00000523261.1
ENST00000380871.4
|
NKX3-1
|
NK3 homeobox 1
|
|
chr10_+_104535994
|
0.018
|
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like
|
|
chr20_-_43589109
|
0.017
|
ENST00000372813.3
|
TOMM34
|
translocase of outer mitochondrial membrane 34
|
|
chr19_+_34663397
|
0.017
|
ENST00000540746.2
ENST00000544216.3
ENST00000433627.5
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr11_+_48002076
|
0.016
|
ENST00000418331.2
ENST00000440289.2
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr19_+_13135386
|
0.016
|
ENST00000360105.4
ENST00000588228.1
ENST00000591028.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr3_+_49726932
|
0.016
|
ENST00000327697.6
ENST00000432042.1
ENST00000454491.1
|
RNF123
|
ring finger protein 123
|
|
chr1_+_89149905
|
0.016
|
ENST00000316005.7
ENST00000370521.3
ENST00000370505.3
|
PKN2
|
protein kinase N2
|
|
chr19_+_40697514
|
0.015
|
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10
|
|
chr17_-_6459768
|
0.015
|
ENST00000421306.3
|
PITPNM3
|
PITPNM family member 3
|
|
chr10_-_119806085
|
0.014
|
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I)
|
|
chr11_+_63606373
|
0.014
|
ENST00000402010.2
ENST00000315032.8
ENST00000377809.4
ENST00000413835.2
ENST00000377810.3
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr7_+_43152191
|
0.014
|
ENST00000395891.2
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr12_-_51477333
|
0.014
|
ENST00000228515.1
ENST00000548206.1
ENST00000546935.1
ENST00000548981.1
|
CSRNP2
|
cysteine-serine-rich nuclear protein 2
|
|
chr4_-_149365827
|
0.013
|
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr4_+_77870856
|
0.013
|
ENST00000264893.6
ENST00000502584.1
ENST00000510641.1
|
SEPT11
|
septin 11
|
|
chr10_-_120101804
|
0.013
|
ENST00000369183.4
ENST00000369172.4
|
FAM204A
|
family with sequence similarity 204, member A
|
|
chr2_-_224467093
|
0.013
|
ENST00000305409.2
|
SCG2
|
secretogranin II
|
|
chr8_+_61591337
|
0.012
|
ENST00000423902.2
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr17_+_57970469
|
0.012
|
ENST00000443572.2
ENST00000406116.3
ENST00000225577.4
ENST00000393021.3
|
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1
|
|
chr11_-_65430251
|
0.012
|
ENST00000534283.1
ENST00000527749.1
ENST00000533187.1
ENST00000525693.1
ENST00000534558.1
ENST00000532879.1
ENST00000532999.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A
|
|
chr14_-_31495569
|
0.012
|
ENST00000357479.5
ENST00000355683.5
|
STRN3
|
striatin, calmodulin binding protein 3
|
|
chr9_-_23821273
|
0.011
|
ENST00000380110.4
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2
|
|
chr15_+_92937058
|
0.011
|
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr18_+_158513
|
0.011
|
ENST00000400266.3
ENST00000580410.1
ENST00000383589.2
ENST00000261601.7
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase)
|
|
chr19_-_10514184
|
0.011
|
ENST00000589629.1
ENST00000222005.2
|
CDC37
|
cell division cycle 37
|
|
chr2_-_169887827
|
0.011
|
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr19_+_11071546
|
0.011
|
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr3_+_29322803
|
0.010
|
ENST00000396583.3
ENST00000383767.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr10_-_98346801
|
0.010
|
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr3_-_21792838
|
0.010
|
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D
|
|
chrX_-_129244655
|
0.009
|
ENST00000335997.7
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr14_+_103243813
|
0.009
|
ENST00000560371.1
ENST00000347662.4
ENST00000392745.2
ENST00000539721.1
ENST00000560463.1
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr5_+_156607829
|
0.009
|
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr3_-_142166904
|
0.008
|
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr6_-_107780751
|
0.008
|
ENST00000453874.2
ENST00000369037.4
ENST00000369031.4
|
PDSS2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2
|
|
chr12_+_5019061
|
0.007
|
ENST00000382545.3
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr1_+_154377669
|
0.007
|
ENST00000368485.3
ENST00000344086.4
|
IL6R
|
interleukin 6 receptor
|
|
chr11_-_73309228
|
0.007
|
ENST00000356467.4
ENST00000064778.4
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr8_-_57123815
|
0.007
|
ENST00000316981.3
ENST00000423799.2
ENST00000429357.2
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr19_-_57352064
|
0.007
|
ENST00000326441.9
ENST00000593695.1
ENST00000599577.1
ENST00000594389.1
ENST00000423103.2
ENST00000598410.1
ENST00000593711.1
ENST00000391708.3
ENST00000221722.5
ENST00000599935.1
|
PEG3
ZIM2
|
paternally expressed 3
zinc finger, imprinted 2
|
|
chr1_-_1624083
|
0.006
|
ENST00000378662.1
ENST00000234800.6
|
SLC35E2B
|
solute carrier family 35, member E2B
|
|
chrX_-_19988382
|
0.006
|
ENST00000356980.3
ENST00000379687.3
ENST00000379682.4
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr20_+_32077880
|
0.005
|
ENST00000342704.6
ENST00000375279.2
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr15_+_59279851
|
0.005
|
ENST00000348370.4
ENST00000434298.1
ENST00000559160.1
|
RNF111
|
ring finger protein 111
|
|
chr3_-_50605077
|
0.004
|
ENST00000426034.1
ENST00000441239.1
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr2_+_173600671
|
0.003
|
ENST00000409036.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr4_-_39979576
|
0.003
|
ENST00000303538.8
ENST00000503396.1
|
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae)
|
|
chr10_+_82213904
|
0.003
|
ENST00000429989.3
|
TSPAN14
|
tetraspanin 14
|
|
chr12_+_2162447
|
0.002
|
ENST00000335762.5
ENST00000399655.1
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr16_+_69599861
|
0.001
|
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr10_+_114709999
|
0.001
|
ENST00000355995.4
ENST00000545257.1
ENST00000543371.1
ENST00000536810.1
ENST00000355717.4
ENST00000538897.1
ENST00000534894.1
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr2_-_39348137
|
0.001
|
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila)
|
|
chr7_+_95401851
|
0.000
|
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1
|
|
chr11_-_128392085
|
0.000
|
ENST00000526145.2
ENST00000531611.1
ENST00000319397.6
ENST00000345075.4
ENST00000535549.1
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1
|
|
chr2_+_178077477
|
0.000
|
ENST00000411529.2
ENST00000435711.1
|
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3
|