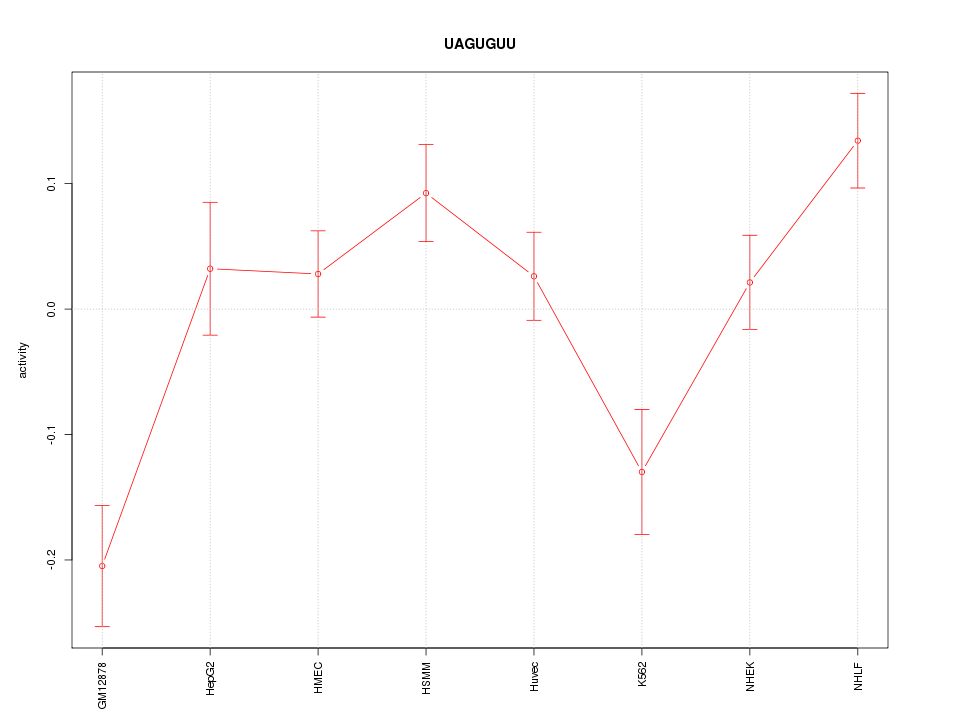
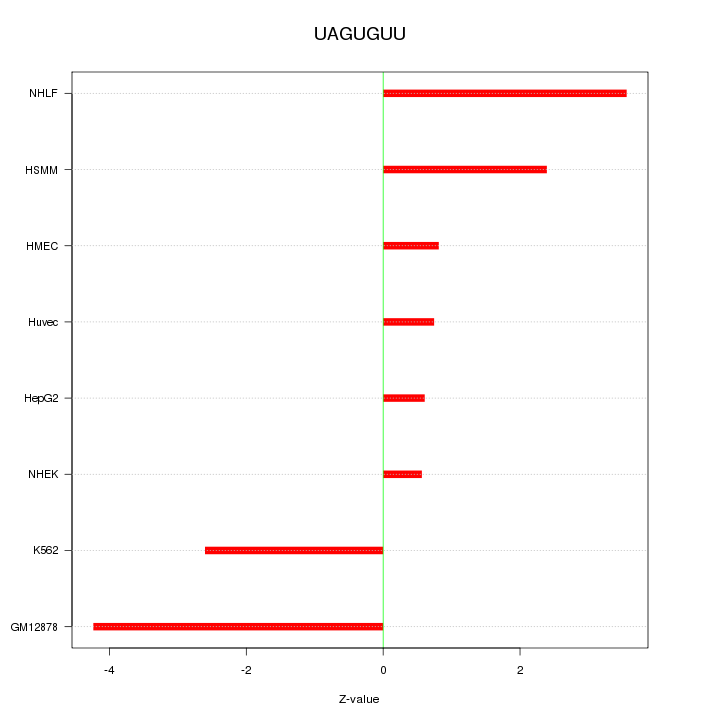
Motif ID: UAGUGUU
Z-value: 2.372

Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) Sertoli cell proliferation(GO:0060011) |
| 0.7 | 3.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.6 | 4.5 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 2.2 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.5 | 2.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.5 | 4.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.4 | 2.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 | 2.8 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.3 | 1.0 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.3 | 1.4 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.2 | 1.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 0.7 | GO:0006740 | glyoxylate cycle(GO:0006097) NADPH regeneration(GO:0006740) |
| 0.2 | 0.7 | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.2 | 1.4 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.2 | 0.6 | GO:0007028 | cytoplasm organization(GO:0007028) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.2 | 0.7 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.7 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.2 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 1.1 | GO:0046349 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 0.3 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 1.9 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) positive regulation of vesicle fusion(GO:0031340) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 1.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 1.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 3.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 1.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 2.9 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.1 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 5.8 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.1 | 0.6 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.1 | 4.1 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 0.7 | GO:0010544 | negative regulation of platelet activation(GO:0010544) negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 0.8 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 4.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 1.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.6 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.1 | 0.2 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.1 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 1.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 3.0 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 2.7 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.5 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.9 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 1.0 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.0 | GO:0051589 | negative regulation of neurotransmitter transport(GO:0051589) |
| 0.0 | 1.2 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.6 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.2 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 3.0 | GO:0007204 | positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 1.2 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0072369 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 2.2 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.1 | GO:0051988 | negative regulation of odontogenesis(GO:0042483) regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.5 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.0 | 0.5 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 2.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 1.7 | GO:0006813 | potassium ion transport(GO:0006813) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.6 | 2.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.4 | 2.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 1.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 0.9 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 1.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 2.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 5.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 2.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 3.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 6.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 1.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 3.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 3.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 4.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.2 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 2.2 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.7 | 4.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.5 | 2.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.4 | 1.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.3 | 3.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 5.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 2.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 1.4 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.3 | 1.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.2 | 0.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 0.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 3.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 3.0 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.2 | 0.6 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.2 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 2.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.2 | 1.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 0.8 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 1.3 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.1 | 1.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 1.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 3.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 2.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.9 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 2.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 5.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.3 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 3.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 1.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 3.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.7 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.2 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.9 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.3 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |