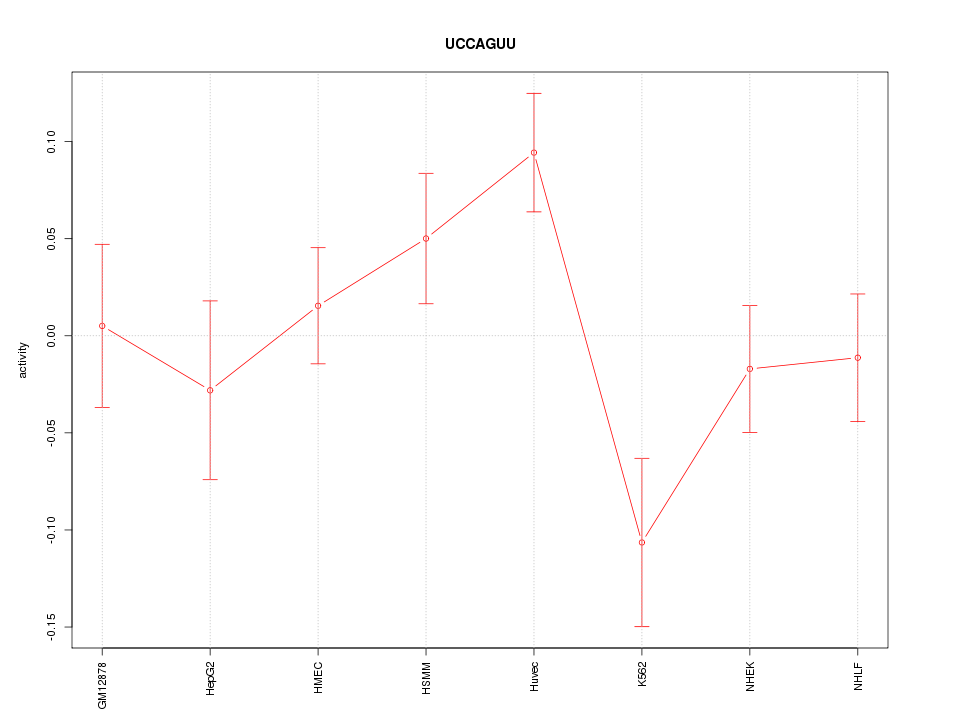
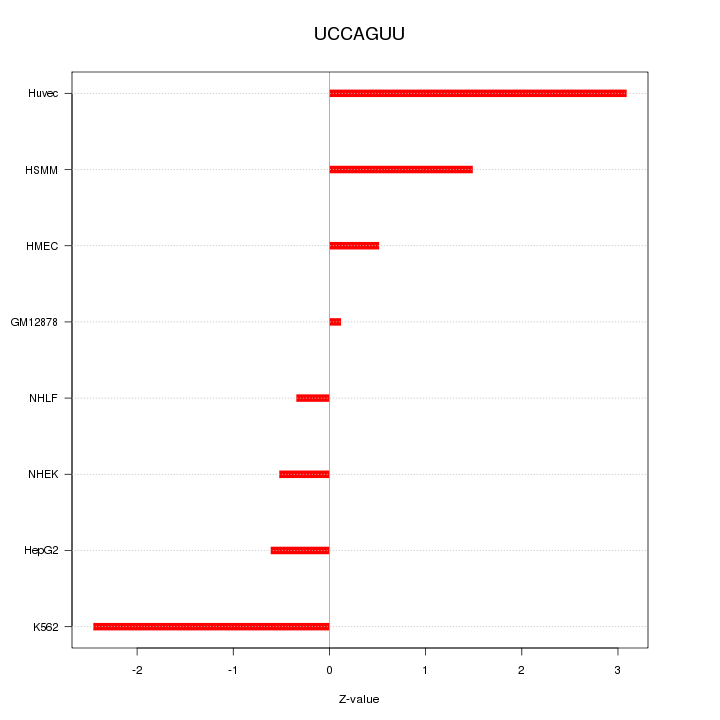
| 0.7 |
2.0 |
GO:0010757 |
chronological cell aging(GO:0001300) arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) negative regulation of plasminogen activation(GO:0010757) negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.6 |
3.2 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.5 |
1.6 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.5 |
1.5 |
GO:0002514 |
B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.5 |
0.9 |
GO:0060535 |
trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.4 |
1.7 |
GO:0006663 |
platelet activating factor biosynthetic process(GO:0006663) positive regulation of vesicle fusion(GO:0031340) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.4 |
1.6 |
GO:0050928 |
negative regulation of positive chemotaxis(GO:0050928) |
| 0.4 |
2.2 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.3 |
1.8 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.3 |
1.7 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.3 |
2.7 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 |
1.0 |
GO:0034959 |
neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 |
0.8 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 |
0.4 |
GO:0003176 |
aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) |
| 0.2 |
0.6 |
GO:0061298 |
progesterone secretion(GO:0042701) retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 |
0.6 |
GO:0046881 |
positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.2 |
0.9 |
GO:0021535 |
cell migration in hindbrain(GO:0021535) |
| 0.2 |
0.8 |
GO:0044854 |
membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) positive regulation of dopamine receptor signaling pathway(GO:0060161) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 0.1 |
0.9 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 |
0.4 |
GO:0045726 |
regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 |
0.1 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 |
0.2 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.1 |
0.9 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
0.5 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
1.2 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 |
0.2 |
GO:0046520 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 |
0.6 |
GO:0032528 |
microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 |
0.1 |
GO:0006679 |
glucosylceramide biosynthetic process(GO:0006679) |
| 0.1 |
0.2 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 |
0.4 |
GO:0032527 |
retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.1 |
0.2 |
GO:0033025 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
0.4 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.1 |
0.3 |
GO:0051051 |
negative regulation of transport(GO:0051051) |
| 0.1 |
0.6 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.1 |
0.1 |
GO:0070672 |
response to interleukin-15(GO:0070672) |
| 0.1 |
0.6 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 |
0.3 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.0 |
0.7 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.5 |
GO:0007351 |
tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 |
0.8 |
GO:0030949 |
positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 |
1.9 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 |
0.3 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 |
0.4 |
GO:0001569 |
patterning of blood vessels(GO:0001569) |
| 0.0 |
0.3 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.2 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 |
0.2 |
GO:0019471 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 |
0.5 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.2 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.3 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
3.2 |
GO:0030048 |
actin filament-based movement(GO:0030048) |
| 0.0 |
0.3 |
GO:0035196 |
production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 |
0.1 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 |
0.4 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
1.2 |
GO:0045768 |
obsolete positive regulation of anti-apoptosis(GO:0045768) |
| 0.0 |
1.0 |
GO:0002208 |
somatic recombination of immunoglobulin genes involved in immune response(GO:0002204) somatic diversification of immunoglobulins involved in immune response(GO:0002208) isotype switching(GO:0045190) |
| 0.0 |
0.3 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 |
0.9 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.0 |
0.3 |
GO:0051602 |
response to electrical stimulus(GO:0051602) |
| 0.0 |
0.1 |
GO:0060744 |
thelarche(GO:0042695) development of secondary female sexual characteristics(GO:0046543) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 |
0.3 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.0 |
0.4 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.1 |
GO:0001999 |
renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) |
| 0.0 |
0.4 |
GO:1903729 |
regulation of establishment of protein localization to plasma membrane(GO:0090003) regulation of protein localization to plasma membrane(GO:1903076) regulation of plasma membrane organization(GO:1903729) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 |
0.1 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
1.7 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.0 |
0.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.2 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.0 |
0.1 |
GO:0097576 |
vacuole fusion(GO:0097576) |
| 0.0 |
1.9 |
GO:0065004 |
protein-DNA complex assembly(GO:0065004) |
| 0.0 |
0.4 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.2 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.2 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.0 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 |
0.1 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 |
0.3 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |