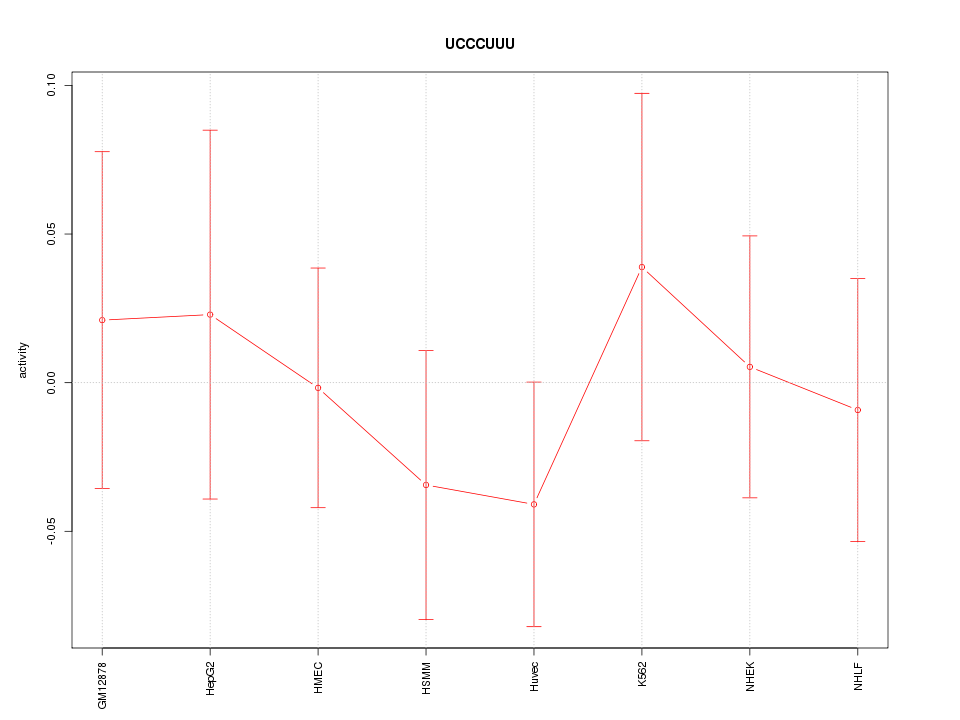
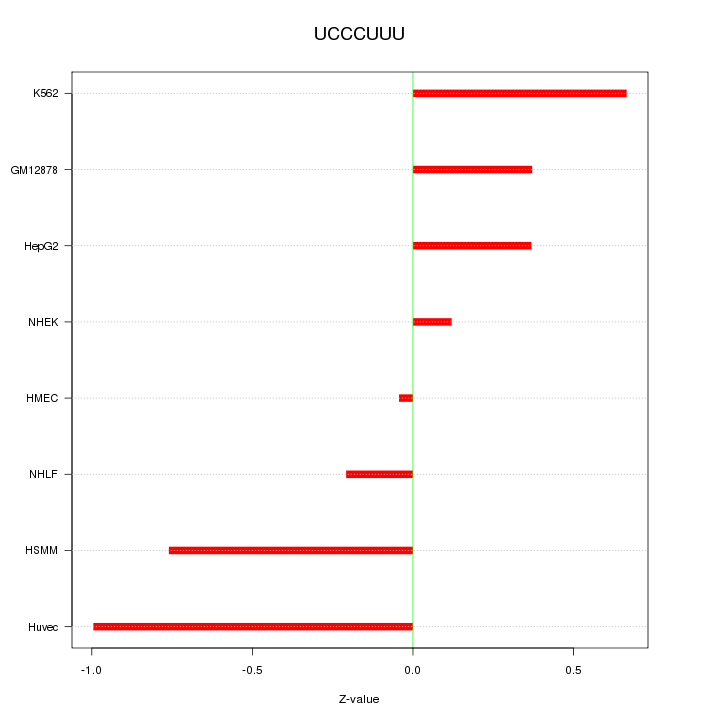
| 0.1 |
0.3 |
GO:1901419 |
regulation of response to alcohol(GO:1901419) |
| 0.1 |
0.3 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
0.3 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.1 |
0.4 |
GO:0051570 |
regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 |
0.2 |
GO:0071539 |
Golgi localization(GO:0051645) protein localization to centrosome(GO:0071539) |
| 0.0 |
0.3 |
GO:0045722 |
response to muscle activity(GO:0014850) positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.2 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 |
0.2 |
GO:0006391 |
transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 |
0.1 |
GO:0046671 |
regulation of nitrogen utilization(GO:0006808) positive regulation of neuron maturation(GO:0014042) nitrogen utilization(GO:0019740) cochlear nucleus development(GO:0021747) negative regulation of cellular pH reduction(GO:0032848) glial cell apoptotic process(GO:0034349) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) positive regulation of cell maturation(GO:1903431) |
| 0.0 |
0.1 |
GO:0042759 |
fatty acid elongation, saturated fatty acid(GO:0019367) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
0.2 |
GO:0022614 |
membrane to membrane docking(GO:0022614) |
| 0.0 |
0.2 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 |
0.1 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.1 |
GO:0071025 |
RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 |
0.1 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.1 |
GO:0032071 |
regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of helicase activity(GO:0051097) |
| 0.0 |
0.1 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
0.0 |
GO:0003148 |
outflow tract septum morphogenesis(GO:0003148) |
| 0.0 |
0.3 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 |
0.1 |
GO:0046619 |
optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.3 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 |
0.2 |
GO:0060487 |
lung epithelial cell differentiation(GO:0060487) |
| 0.0 |
0.1 |
GO:0031062 |
positive regulation of histone methylation(GO:0031062) |
| 0.0 |
0.2 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.1 |
GO:0072201 |
negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 |
0.2 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.1 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.0 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.1 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.1 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.1 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.0 |
GO:0002384 |
hepatic immune response(GO:0002384) |
| 0.0 |
0.3 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.2 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.3 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |