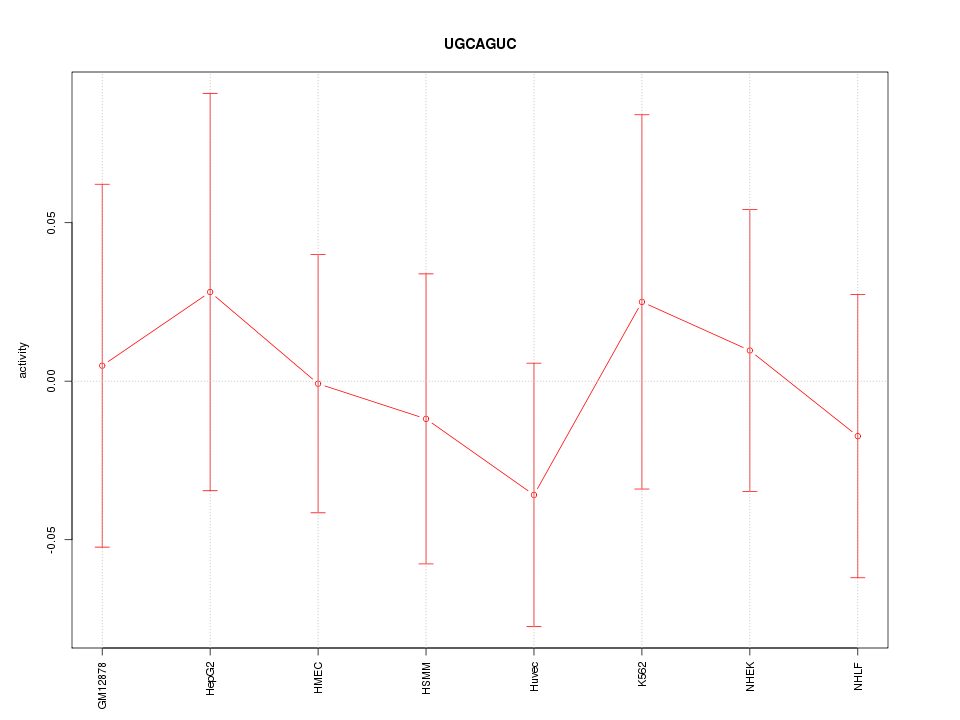
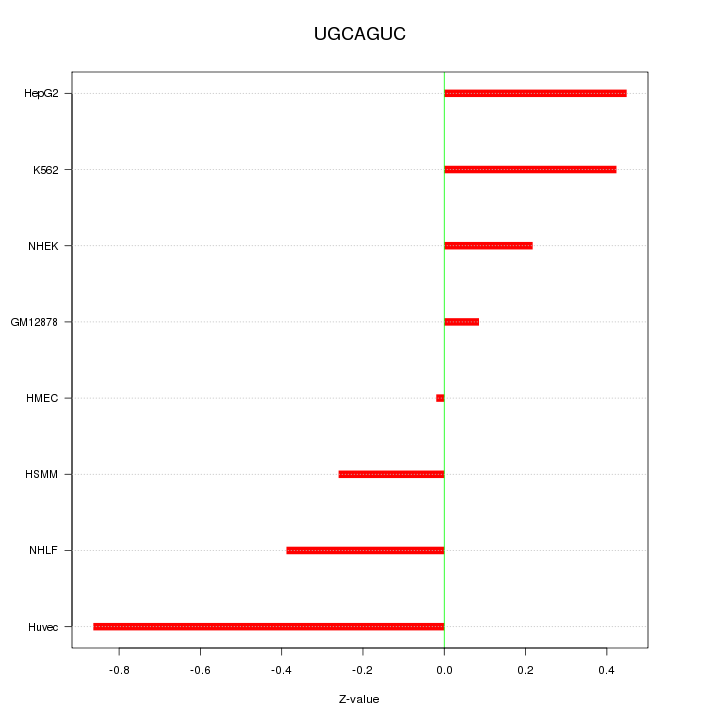
|
chr6_-_119670919
|
0.274
|
ENST00000368468.3
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr4_-_73434498
|
0.226
|
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr1_+_39456895
|
0.203
|
ENST00000432648.3
ENST00000446189.2
ENST00000372984.4
|
AKIRIN1
|
akirin 1
|
|
chr18_+_55102917
|
0.190
|
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2
|
|
chr12_-_48398104
|
0.174
|
ENST00000337299.6
ENST00000380518.3
|
COL2A1
|
collagen, type II, alpha 1
|
|
chr20_-_14318248
|
0.166
|
ENST00000378053.3
ENST00000341420.4
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr10_+_88516396
|
0.163
|
ENST00000372037.3
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr2_-_11484710
|
0.147
|
ENST00000315872.6
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2
|
|
chr5_+_167718604
|
0.144
|
ENST00000265293.4
|
WWC1
|
WW and C2 domain containing 1
|
|
chr11_-_70507901
|
0.138
|
ENST00000449833.2
ENST00000357171.3
ENST00000449116.2
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr10_-_62149433
|
0.136
|
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_+_48189612
|
0.133
|
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9
|
|
chr1_+_11751748
|
0.132
|
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein
|
|
chr16_-_73082274
|
0.117
|
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr19_+_35759824
|
0.115
|
ENST00000343550.5
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr2_-_148778258
|
0.108
|
ENST00000392857.5
ENST00000457954.1
ENST00000392858.1
ENST00000542387.1
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr11_+_125496124
|
0.104
|
ENST00000533778.2
ENST00000534070.1
|
CHEK1
|
checkpoint kinase 1
|
|
chr21_+_22370608
|
0.102
|
ENST00000400546.1
|
NCAM2
|
neural cell adhesion molecule 2
|
|
chr6_-_16761678
|
0.101
|
ENST00000244769.4
ENST00000436367.1
|
ATXN1
|
ataxin 1
|
|
chr16_-_56459354
|
0.099
|
ENST00000290649.5
|
AMFR
|
autocrine motility factor receptor, E3 ubiquitin protein ligase
|
|
chr6_+_161412759
|
0.099
|
ENST00000366919.2
ENST00000392142.4
ENST00000366920.2
ENST00000348824.7
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr10_-_47213626
|
0.097
|
ENST00000452145.2
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10
|
|
chr2_-_2334888
|
0.094
|
ENST00000428368.2
ENST00000399161.2
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr10_-_105615164
|
0.093
|
ENST00000355946.2
ENST00000369774.4
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr2_+_228336849
|
0.092
|
ENST00000409979.2
ENST00000310078.8
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr6_-_30585009
|
0.091
|
ENST00000376511.2
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10
|
|
chr3_-_53880401
|
0.084
|
ENST00000315251.6
|
CHDH
|
choline dehydrogenase
|
|
chr4_-_99851766
|
0.083
|
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr16_+_50187556
|
0.082
|
ENST00000561678.1
ENST00000357464.3
|
PAPD5
|
PAP associated domain containing 5
|
|
chr3_-_129407535
|
0.080
|
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr15_-_83316254
|
0.079
|
ENST00000567678.1
ENST00000450751.2
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr12_+_62654119
|
0.079
|
ENST00000353364.3
ENST00000549523.1
ENST00000280377.5
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr7_+_28452130
|
0.079
|
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr15_-_34628951
|
0.078
|
ENST00000397707.2
ENST00000560611.1
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6
|
|
chr14_+_93651358
|
0.075
|
ENST00000415050.2
|
TMEM251
|
transmembrane protein 251
|
|
chr21_+_40177143
|
0.070
|
ENST00000360214.3
|
ETS2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2
|
|
chr1_+_167190066
|
0.070
|
ENST00000367866.2
ENST00000429375.2
ENST00000452019.1
ENST00000420254.3
ENST00000541643.3
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr2_+_48010221
|
0.069
|
ENST00000234420.5
|
MSH6
|
mutS homolog 6
|
|
chr12_+_862089
|
0.067
|
ENST00000315939.6
ENST00000537687.1
ENST00000447667.2
|
WNK1
|
WNK lysine deficient protein kinase 1
|
|
chr2_+_118846008
|
0.067
|
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2
|
|
chr1_+_87794150
|
0.065
|
ENST00000370544.5
|
LMO4
|
LIM domain only 4
|
|
chrX_-_110513703
|
0.065
|
ENST00000324068.1
|
CAPN6
|
calpain 6
|
|
chr1_+_36348790
|
0.065
|
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1
|
|
chr14_+_103243813
|
0.064
|
ENST00000560371.1
ENST00000347662.4
ENST00000392745.2
ENST00000539721.1
ENST00000560463.1
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr16_-_12009735
|
0.064
|
ENST00000439887.2
ENST00000434724.2
|
GSPT1
|
G1 to S phase transition 1
|
|
chr19_-_46476791
|
0.063
|
ENST00000263257.5
|
NOVA2
|
neuro-oncological ventral antigen 2
|
|
chr11_-_134281812
|
0.062
|
ENST00000392580.1
ENST00000312527.4
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P)
|
|
chr11_-_72504637
|
0.061
|
ENST00000359373.5
ENST00000536377.1
|
ARAP1
STARD10
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
StAR-related lipid transfer (START) domain containing 10
|
|
chr10_-_94003003
|
0.059
|
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr9_+_112403088
|
0.057
|
ENST00000448454.2
|
PALM2
|
paralemmin 2
|
|
chr12_+_110906169
|
0.056
|
ENST00000377673.5
|
FAM216A
|
family with sequence similarity 216, member A
|
|
chr20_+_62795827
|
0.055
|
ENST00000328439.1
ENST00000536311.1
|
MYT1
|
myelin transcription factor 1
|
|
chr5_+_122110691
|
0.052
|
ENST00000379516.2
ENST00000505934.1
ENST00000514949.1
|
SNX2
|
sorting nexin 2
|
|
chr2_+_191273052
|
0.051
|
ENST00000417958.1
ENST00000432036.1
ENST00000392328.1
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr15_+_33603147
|
0.050
|
ENST00000415757.3
ENST00000389232.4
|
RYR3
|
ryanodine receptor 3
|
|
chr19_-_12834739
|
0.050
|
ENST00000589337.1
ENST00000425528.1
ENST00000441499.1
ENST00000588216.1
|
TNPO2
|
transportin 2
|
|
chr9_-_37034028
|
0.050
|
ENST00000520281.1
ENST00000446742.1
ENST00000522003.1
ENST00000523145.1
ENST00000414447.1
ENST00000377847.2
ENST00000377853.2
ENST00000377852.2
ENST00000523241.1
ENST00000520154.1
ENST00000358127.4
|
PAX5
|
paired box 5
|
|
chr10_+_89622870
|
0.048
|
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog
|
|
chr2_+_166326157
|
0.045
|
ENST00000421875.1
ENST00000314499.7
ENST00000409664.1
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr3_-_164913777
|
0.042
|
ENST00000475390.1
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr12_+_104850740
|
0.041
|
ENST00000547956.1
ENST00000549260.1
ENST00000303694.5
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr3_-_21792838
|
0.040
|
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D
|
|
chr1_-_217262969
|
0.039
|
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr7_+_101928380
|
0.039
|
ENST00000536178.1
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr5_-_32444828
|
0.039
|
ENST00000265069.8
|
ZFR
|
zinc finger RNA binding protein
|
|
chr1_+_36396677
|
0.039
|
ENST00000373191.4
ENST00000397828.2
|
AGO3
|
argonaute RISC catalytic component 3
|
|
chr5_+_36876833
|
0.038
|
ENST00000282516.8
ENST00000448238.2
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr11_+_33278811
|
0.038
|
ENST00000303296.4
ENST00000379016.3
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr3_+_137906109
|
0.037
|
ENST00000481646.1
ENST00000469044.1
ENST00000491704.1
ENST00000461600.1
|
ARMC8
|
armadillo repeat containing 8
|
|
chr1_+_233086326
|
0.036
|
ENST00000366628.5
ENST00000366627.4
|
NTPCR
|
nucleoside-triphosphatase, cancer-related
|
|
chr5_-_133561752
|
0.036
|
ENST00000481195.1
ENST00000519718.1
|
PPP2CA
CTD-2410N18.5
|
protein phosphatase 2, catalytic subunit, alpha isozyme
S-phase kinase-associated protein 1
|
|
chr12_-_16761007
|
0.034
|
ENST00000354662.1
ENST00000441439.2
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr17_-_38020392
|
0.034
|
ENST00000346872.3
ENST00000439167.2
ENST00000377945.3
ENST00000394189.2
ENST00000377944.3
ENST00000377958.2
ENST00000535189.1
ENST00000377952.2
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos)
|
|
chr16_-_46865047
|
0.034
|
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87
|
|
chr3_-_98620500
|
0.034
|
ENST00000326840.6
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr20_+_3451650
|
0.032
|
ENST00000262919.5
|
ATRN
|
attractin
|
|
chr2_+_28974668
|
0.031
|
ENST00000296122.6
ENST00000395366.2
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme
|
|
chr10_-_5855350
|
0.031
|
ENST00000456041.1
ENST00000380181.3
ENST00000418688.1
ENST00000380132.4
ENST00000609712.1
ENST00000380191.4
|
GDI2
|
GDP dissociation inhibitor 2
|
|
chr2_-_154335300
|
0.031
|
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr15_+_77287426
|
0.031
|
ENST00000558012.1
ENST00000267939.5
ENST00000379595.3
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr7_-_105517021
|
0.030
|
ENST00000318724.4
ENST00000419735.3
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr7_+_119913688
|
0.030
|
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chrX_+_107683096
|
0.030
|
ENST00000328300.6
ENST00000361603.2
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr14_+_71108460
|
0.028
|
ENST00000256367.2
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr7_+_94139105
|
0.028
|
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1
|
|
chr2_+_149632783
|
0.026
|
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C
|
|
chr14_-_21737610
|
0.026
|
ENST00000320084.7
ENST00000449098.1
ENST00000336053.6
|
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2)
|
|
chr1_-_53793584
|
0.024
|
ENST00000354412.3
ENST00000347547.2
ENST00000306052.6
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr1_+_237205476
|
0.023
|
ENST00000366574.2
|
RYR2
|
ryanodine receptor 2 (cardiac)
|
|
chr7_-_32931387
|
0.022
|
ENST00000304056.4
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr17_-_42580738
|
0.022
|
ENST00000585614.1
ENST00000591680.1
ENST00000434000.1
ENST00000588554.1
ENST00000592154.1
|
GPATCH8
|
G patch domain containing 8
|
|
chr2_+_46926048
|
0.022
|
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr1_-_78444776
|
0.021
|
ENST00000370767.1
ENST00000421641.1
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr1_+_28052456
|
0.021
|
ENST00000373954.6
ENST00000419687.2
|
FAM76A
|
family with sequence similarity 76, member A
|
|
chr3_-_15106747
|
0.019
|
ENST00000449354.2
ENST00000444840.2
ENST00000253686.2
|
MRPS25
|
mitochondrial ribosomal protein S25
|
|
chr4_-_76598296
|
0.017
|
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr5_+_149109825
|
0.016
|
ENST00000360453.4
ENST00000394320.3
ENST00000309241.5
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_-_35658736
|
0.015
|
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr18_+_63418068
|
0.015
|
ENST00000397968.2
|
CDH7
|
cadherin 7, type 2
|
|
chr11_-_132813566
|
0.014
|
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr2_+_204192942
|
0.013
|
ENST00000295851.5
ENST00000261017.5
|
ABI2
|
abl-interactor 2
|
|
chr14_+_94492674
|
0.012
|
ENST00000203664.5
ENST00000553723.1
|
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2
|
|
chr4_-_42659102
|
0.010
|
ENST00000264449.10
ENST00000510289.1
ENST00000381668.5
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr10_+_97803151
|
0.009
|
ENST00000403870.3
ENST00000265992.5
ENST00000465148.2
ENST00000534974.1
|
CCNJ
|
cyclin J
|
|
chr10_-_120514720
|
0.008
|
ENST00000369151.3
ENST00000340214.4
|
CACUL1
|
CDK2-associated, cullin domain 1
|
|
chr1_-_70671216
|
0.007
|
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr3_+_121903181
|
0.007
|
ENST00000498619.1
|
CASR
|
calcium-sensing receptor
|
|
chr3_-_12200851
|
0.006
|
ENST00000287814.4
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr1_+_171454659
|
0.005
|
ENST00000367742.3
ENST00000338920.4
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr17_-_73775839
|
0.005
|
ENST00000592643.1
ENST00000591890.1
ENST00000587171.1
ENST00000254810.4
ENST00000589599.1
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr2_-_69870835
|
0.005
|
ENST00000409085.4
ENST00000406297.3
|
AAK1
|
AP2 associated kinase 1
|
|
chr3_+_126707437
|
0.004
|
ENST00000393409.2
ENST00000251772.4
|
PLXNA1
|
plexin A1
|
|
chr14_-_55878538
|
0.004
|
ENST00000247178.5
|
ATG14
|
autophagy related 14
|
|
chr9_-_131790550
|
0.002
|
ENST00000372554.4
ENST00000372564.3
|
SH3GLB2
|
SH3-domain GRB2-like endophilin B2
|
|
chr6_+_142468361
|
0.000
|
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1
|