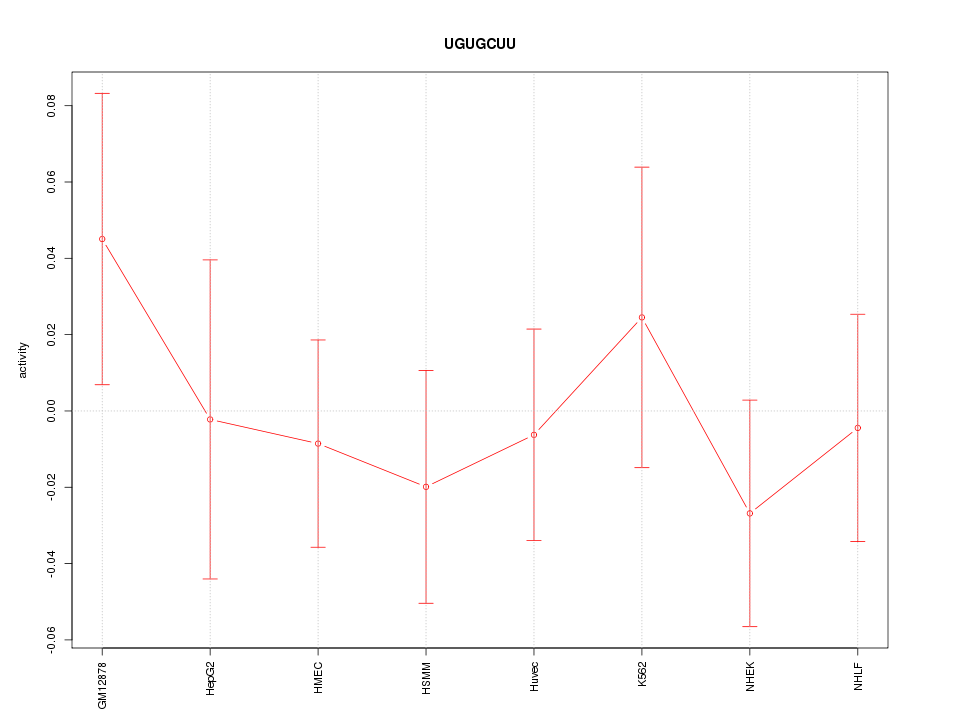
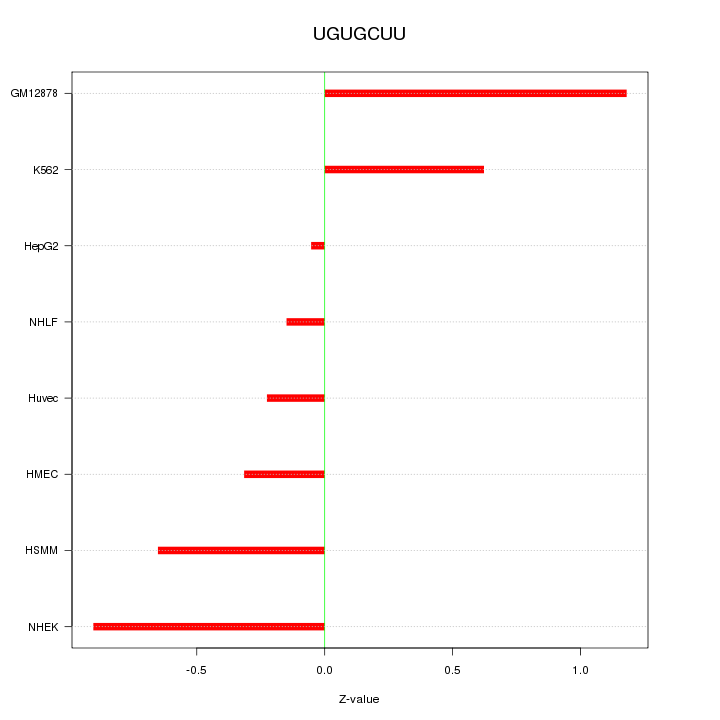
| 0.1 |
0.3 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
0.4 |
GO:0048935 |
peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.3 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.1 |
0.3 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 |
0.3 |
GO:0007109 |
obsolete cytokinesis, completion of separation(GO:0007109) protein hexamerization(GO:0034214) |
| 0.1 |
0.3 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 |
0.3 |
GO:0032776 |
DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.1 |
0.3 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.1 |
0.5 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 |
0.2 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 |
0.4 |
GO:0021903 |
rostrocaudal neural tube patterning(GO:0021903) |
| 0.1 |
0.2 |
GO:2000078 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) regulation of type B pancreatic cell development(GO:2000074) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 |
0.3 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.5 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
0.3 |
GO:0032916 |
transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 |
0.2 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 |
0.1 |
GO:0008050 |
female courtship behavior(GO:0008050) |
| 0.0 |
0.2 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.0 |
0.3 |
GO:0009082 |
branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.0 |
0.3 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 |
0.1 |
GO:0007028 |
cytoplasm organization(GO:0007028) |
| 0.0 |
0.1 |
GO:0034625 |
fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.3 |
GO:0046349 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 |
0.3 |
GO:0019264 |
L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.0 |
0.5 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.3 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.2 |
GO:0045767 |
obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 |
0.1 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.1 |
GO:0014889 |
muscle atrophy(GO:0014889) |
| 0.0 |
0.2 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.1 |
GO:0055005 |
optic placode formation involved in camera-type eye formation(GO:0046619) ventricular cardiac myofibril assembly(GO:0055005) embryonic retina morphogenesis in camera-type eye(GO:0060059) regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 |
0.2 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.0 |
0.2 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.1 |
GO:0072202 |
cell differentiation involved in metanephros development(GO:0072202) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.3 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.1 |
GO:1904036 |
negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.1 |
GO:0050685 |
positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 |
0.1 |
GO:0032933 |
SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.2 |
GO:0007090 |
obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 |
0.4 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.2 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.2 |
GO:0043306 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.2 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) regulation of phospholipase activity(GO:0010517) positive regulation of phospholipase activity(GO:0010518) positive regulation of phospholipase C activity(GO:0010863) regulation of lipase activity(GO:0060191) positive regulation of lipase activity(GO:0060193) regulation of phospholipase C activity(GO:1900274) |
| 0.0 |
0.4 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.1 |
GO:1903307 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) positive regulation of regulated secretory pathway(GO:1903307) |
| 0.0 |
0.3 |
GO:0006448 |
regulation of translational elongation(GO:0006448) |
| 0.0 |
0.3 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.3 |
GO:0051057 |
positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 |
0.1 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.0 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.0 |
0.4 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 0.0 |
0.2 |
GO:0042354 |
fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 |
0.0 |
GO:0060124 |
positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 |
0.3 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.0 |
0.1 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.1 |
GO:0006772 |
thiamine metabolic process(GO:0006772) |
| 0.0 |
0.1 |
GO:0060992 |
response to fungicide(GO:0060992) |
| 0.0 |
0.2 |
GO:0006906 |
vesicle fusion(GO:0006906) |
| 0.0 |
0.1 |
GO:0016265 |
obsolete death(GO:0016265) |
| 0.0 |
0.1 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.0 |
0.0 |
GO:0097576 |
autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.0 |
0.1 |
GO:0032075 |
positive regulation of nuclease activity(GO:0032075) |
| 0.0 |
0.3 |
GO:0002076 |
osteoblast development(GO:0002076) |
| 0.0 |
0.5 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 |
0.2 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.2 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.4 |
GO:0001508 |
action potential(GO:0001508) |
| 0.0 |
0.7 |
GO:0061025 |
membrane fusion(GO:0061025) |
| 0.0 |
0.2 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.2 |
GO:0048260 |
positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 |
0.0 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 |
0.0 |
GO:0060750 |
epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 |
0.1 |
GO:0044060 |
regulation of endocrine process(GO:0044060) |
| 0.0 |
0.3 |
GO:0051925 |
obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 |
0.1 |
GO:0000012 |
single strand break repair(GO:0000012) |